Key Points
-
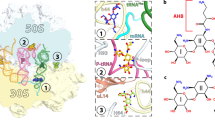
A large proportion of clinically useful antibiotics exert their antimicrobial effects by blocking protein synthesis on the ribosome. The bacterial ribosome is a ribonucleoprotein complex of about 2.5 million Daltons, and is composed of two subunits that are named after their sedimentation values of 30S and 50S.
-
The molecular details of the ribosome have recently been determined by X-ray crystallography. Different organisms have been used as the source of ribosomal particles for crystallization.
-
Well resolved structures have been obtained for the 30S subunit and the intact ribosome from the bacterium Thermus thermophilus. The best resolved structures for the 50S subunit come from the bacterium Deinococcus radiodurans and the archaeon Haloarcula marismortui.
-
These crystal structures reveal the molecular details of the antibiotic-binding sites. Furthermore, they explain many earlier observations from biochemical and genetic studies including: how drugs exercise their inhibitory effects; how some drugs in combination enhance or impede each other's binding; and how alterations to ribosomal components confer resistance.
-
The antibiotic-binding sites are located within functionally important structures in the ribosomal RNA (rRNA). Antibiotic resistance is often conferred by base substitution or methylation at these sites in the rRNA. However, resistance can also be conferred by mutations in ribosomal proteins that influence these rRNA structures.
-
Resistance can be counteracted by equipping current antibiotics with new chemical substituents that improve their binding. Perhaps even greater potential, which is presently unrealized, lies in the rational design of novel compounds that target unexploited sites within the ribosome structure.
Abstract
Many clinically useful antibiotics exert their antimicrobial effects by blocking protein synthesis on the bacterial ribosome. The structure of the ribosome has recently been determined by X-ray crystallography, revealing the molecular details of the antibiotic-binding sites. The crystal data explain many earlier biochemical and genetic observations, including how drugs exercise their inhibitory effects, how some drugs in combination enhance or impede each other's binding, and how alterations to ribosomal components confer resistance. The crystal structures also provide insight as to how existing drugs might be derivatized (or novel drugs created) to improve binding and circumvent resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vázquez, D. Inhibitors of Protein Biosynthesis (Springer–Verlag, Berlin, 1979).
Gale, E. F., Cundliffe, E., Reynolds, P. E., Richmond, M. H. & Waring, M. J. The Molecular Basis of Antibiotic Action (John Wiley and Sons, London, 1981).
Spahn, C. M. & Prescott, C. D. Throwing a spanner in the works: antibiotics and the translation apparatus. J. Mol. Med. 74, 423–439 (1996).
Mankin, A. S. Ribosomal antibiotics. Mol. Biol. 35, 509–520 (2001).
Lake, J. A. Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J. Mol. Biol. 105, 131–139 (1976).
Tischendorf, G. W., Zeichhardt, H. & Stoffler, G. Architecture of the Escherichia coli ribosome as determined by immune electron microscopy. Proc. Natl Acad. Sci. USA 72, 4820–4824 (1975).
Boublik, M., Hellmann, W. & Kleinschmidt, A. K. Size and structure of Escherichia coli ribosomes by electron microscopy. Cytobiologie 14, 293–300 (1977).
Pape, T., Stark, H., Matadeen, R., Orlova, E. V. & van Heel, M. in The Ribosome: Structure, Function, Antibiotics and Cellular Interactions (eds Garrett, R. A. et al.) 37–44 (American Society for Microbiology, Washington DC, 2000).
Agrawal, R. K. & Frank, J. Structural studies of the translational apparatus. Curr. Opin. Struct. Biol. 9, 215–221 (1999).
Frank, J. et al. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett. 579, 959–962 (2005).
Valle, M. et al. Visualizing tmRNA entry into a stalled ribosome. Science 300, 127–130 (2003).
Valle, M. et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nature Struct. Biol. 10, 899–906 (2003). References 11 and 12 illustrate the power of the cryo-EM technique for viewing the ribosome at isolated steps in the translation process.
Noller, H. F., Hoffarth, V. & Zimniak, L. Unusual resistance of peptidyl transferase to protein extraction procedures. 256, 1416–1419 (1992).
Green, R., Switzer, C. & Noller, H. F. Ribosome-catalyzed peptide-bond formation with an A-site substrate covalently linked to 23S ribosomal RNA. Science 280, 286–289 (1998).
Zhang, B. & Cech, T. R. Peptide bond formation by in vitro selected ribozymes. Nature 390, 96–100 (1997).
Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000). The first structural data strongly indicating that peptide-bond formation is RNA-catalysed, and implicating 23S rRNA nucleotide A2451 in this process.
Bashan, A. et al. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11, 91–102 (2003).
Moore, P. B. & Steitz, T. A. The structural basis of large ribosomal subunit function. Annu. Rev. Biochem. 72, 813–850 (2003).
Sievers, A., Beringer, M., Rodnina, M. V. & Wolfenden, R. The ribosome as an entropy trap. Proc. Natl Acad. Sci. USA 101, 7897–7901 (2004).
Youngman, E. M., Brunelle, J. L., Kochaniak, A. B. & Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 (2004).
Erlacher, M. D. et al. Chemical engineering of the peptidyl transferase center reveals an important role of the 2′-hydroxyl group of A2451. Nucleic Acids Res. 33, 1618–1627 (2005). A cleverly designed biochemical/molecular genetic study of peptide-bond formation and the role of the nucleotide A2451 ribose. This article's introduction concisely sums up previous research and discussions on this topic.
Yonath, A., Mussig, J. & Wittmann, H. G. Parameters for crystal growth of ribosomal subunits. J. Cell. Biochem. 19, 145–155 (1982).
Trakhanov, S. et al. Preliminary X-ray investigation of 70 S ribosome crystals from Thermus thermophilus. J. Mol. Biol. 209, 327–328 (1989).
von Bohlen, K. et al. Characterization and preliminary attempts for derivatization of crystals of large ribosomal subunits from Haloarcula marismortui diffracting to 3 Å resolution. J. Mol. Biol. 222, 11–15 (1991).
Yusupov, M. M., Garber, M. B., Vasiliev, V. D. & Spirin, A. S. Thermus thermophilus ribosomes for crystallographic studies. Biochimie 73, 887–897 (1991).
Liljas, A. Structural aspects of protein synthesis (World Scientific, Singapore, 2004).
Wimberly, B. T. et al. Structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000).
Schluenzen, F. et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623 (2000).
Yusupov, M. M. et al. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 (2001). Describes the crystal structures of both ribosomal subunits in functional complexes, and reveals essential details of tRNA interactions and how the ribosome works.
Yusupova, G. Z., Yusupov, M. M., Cate, J. H. & Noller, H. F. The path of messenger RNA through the ribosome. Cell 106, 233–241 (2001).
Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Harms, J. et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688 (2001).
Vila-Sanjurjo, A. et al. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc. Natl Acad. Sci. USA 100, 8682–8687 (2003). First structure of E. coli ribosomes, which had previously been thought to be refractory to crystallographic analysis. Presently at low resolution, but more highly resolved data are on the way.
Klein, D. J., Moore, P. B. & Steitz, T. A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340, 141–177 (2004).
Semrad, K., Green, R. & Schroeder, R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA 10, 1855–1860 (2004).
Noller, H. F. The driving force for molecular evolution of translation. RNA 10, 1833–1837 (2004).
Takyar, S., Hickerson, R. P. & Noller, H. F. mRNA helicase activity of the ribosome. Cell 120, 49–58 (2005).
Gabashvili, I. S. et al. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 8, 181–188 (2001).
Tenson, T. & Ehrenberg, M. Regulatory nascent peptides in the ribosomal tunnel. Cell 108, 591–594 (2002).
Berisio, R. et al. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nature Struct. Biol. 10, 366–370 (2003).
Nakatogawa, H., Murakami, A. & Ito, K. Control of SecA and SecM translation by protein secretion. Curr. Opin. Microbiol. 7, 145–150 (2004).
Tu, D., Blaha, G., Moore, P. B. & Steitz, T. A. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121, 257–270 (2005). An important refinement in the use of Haloarcula subunits for antibiotic binding studies. Also sheds light on the mechanisms of nucleotide A2058 and r-protein L22 mutations in macrolide resistance.
Ferbitz, L. et al. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590–596 (2004).
Fourmy, D., Recht, M. I., Blanchard, S. C. & Puglisi, J. D. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274, 1367–1371 (1996).
Yoshizawa, S., Fourmy, D. & Puglisi, J. D. Structural origins of gentamicin antibiotic action. EMBO J. 17, 6437–6448 (1998).
Carter, A. P. et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407, 340–348 (2000). References 16, 46 and 57 are examples of initial ground-breaking studies by three different research groups that revealed the molecular details of ribosomal subunits and antibiotic interactions. For all these studies, the readers are referred to the atomic coordinates in the databases ( Tables 2 and 3).
Vicens, Q. & Westhof, E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure (Camb.) 9, 647–658 (2001).
Vicens, Q. & Westhof, E. Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A site oligonucleotide. J. Mol. Biol. 326, 1175–1188 (2003).
Hermann, T. Drugs targeting the ribosome. Curr. Opin. Struct. Biol. 15, 355–366 (2005).
Rodnina, M. V. et al. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl Acad. Sci. USA 96, 9586–9590 (1999).
Brandi, L. et al. The translation initiation functions of IF2: targets for thiostrepton inhibition. J. Mol. Biol. 335, 881–894 (2004).
Bowen, W. S., Van Dyke, N., Murgola, E. J., Lodmell, J. S. & Hill, W. E. Interaction of thiostrepton and elongation factor-G with the ribosomal protein L11-binding domain. J. Biol. Chem. 280, 2934–2943 (2005).
McNicholas, P. M. et al. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both Gram-positive and Gram-negative bacteria. Antimicrob. Agents Chemother. 44, 1121–1126 (2000).
Belova, L., Tenson, T., Xiong, L., McNicholas, P. M. & Mankin, A. S. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc. Natl Acad. Sci. USA 98, 3726–3731 (2001).
Kofoed, C. B. & Vester, B. Interaction of avilamycin with ribosomes and resistance caused by mutations in 23S rRNA. Antimicrob. Agents Chemother. 46, 3339–3342 (2002).
Treede, I. et al. The avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA at the guanosine 2535 base and the uridine 2479 ribose. Mol. Microbiol. 49, 309–318 (2003).
Schlunzen, F. et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413, 814–821 (2001).
Hansen, J. L. et al. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10, 117–128 (2002).
Hansen, J. L., Moore, P. B. & Steitz, T. A. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330, 1061–1075 (2003).
Harms, J. M., Schlunzen, F., Fucini, P., Bartels, H. & Yonath, A. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2, 4 (2004).
Pringle, M., Poehlsgaard, J., Vester, B. & Long, K. S. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 54, 1295–1306 (2004).
Schlunzen, F., Pyetan, E., Fucini, P., Yonath, A. & Harms, J. M. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol. Microbiol. 54, 1287–1294 (2004).
Kloss, P., Xiong, L., Shinabarger, D. L. & Mankin, A. S. Resistance mutations in 23 S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294, 93–101 (1999).
Thompson, J., O'Connor, M., Mills, J. A. & Dahlberg, A. E. The protein synthesis inhibitors, oxazolidinones and chloramphenicol, cause extensive translational inaccuracy in vivo. J. Mol. Biol. 322, 273–279 (2002).
Yoshizawa, S., Fourmy, D. & Puglisi, J. D. Recognition of the codon-anticodon helix by ribosomal RNA. Science 285, 1722–1725 (1999). First conclusive experimental data that link the 16S rRNA bases A1492 and A1493 with the decoding process.
Ogle, J. M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001). Crystal study that confirms the involvement of A1492 and A1493 in the decoding process and links their movement with other conformational changes in the 30S ribosomal subunit. See also reference 67 for a review.
Ogle, J. M., Carter, A. P. & Ramakrishnan, V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28, 259–266 (2003).
Piepenburg, O. et al. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry 39, 1734–1738 (2000).
Cochella, L. & Green, R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308, 1178–1180 (2005).
Moazed, D. & Noller, H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327, 389–394 (1987).
Pfister, P. et al. Mutagenesis of 16S rRNA C1409–G1491 base-pair differentiates between 6′-OH and 6′-NH3+ aminoglycosides. J. Mol. Biol. 346, 467–475 (2005).
Gregory, S. T., Carr, J. F. & Dahlberg, A. E. A mutation in the decoding center of Thermus thermophilus 16S rRNA suggests a novel mechanism of streptomycin resistance. J. Bacteriol. 187, 2200–2202 (2005).
Thompson, J., Skeggs, P. A. & Cundliffe, E. Methylation of 16S ribosomal RNA and resistance to the aminoglycoside antibiotics gentamicin and kanamycin determined by DNA from the gentamicin-producer, Micromonospora purpurea. Mol. Gen. Genet. 201, 168–173 (1985).
Beauclerk, A. A. & Cundliffe, E. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J. Mol. Biol. 193, 661–671 (1987).
Douthwaite, S., Fourmy, D. & Yoshizawa, S. in Fine-tuning of RNA Functions by Modification and Editing (ed. Grosjean, H.) 287–309 (Springer–Verlag, New York, 2005).
Gong, F. & Yanofsky, C. Instruction of translating ribosome by nascent peptide. Science 297, 1864–1867 (2002).
Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585 (1995).
Cundliffe, E. in The Ribosome: Structure, Function and Evolution (eds Hill, W. E. et al.) 479–490 (American Society for Microbiology, Washington DC, 1990).
Bryskier, A. J., Butzler, J. P., Neu, H. C. & Tulkens, P. M. Macrolides: Chemistry, Pharmacology and Clinical Uses (Arnette Blackwell, Paris, 1993).
Schönfeld, W. & Kirst, H. A. (eds) Macrolide Antibiotics (Birkhäuser, Basel, 2002).
Tenson, T., Lovmar, M. & Ehrenberg, M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330, 1005–1014 (2003).
Weisblum, B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39, 797–805 (1995).
Tenson, T. & Mankin, A. S. Short peptides conferring resistance to macrolide antibiotics. Peptides 22, 1661–1668 (2001).
Poulsen, S. M., Kofoed, C. & Vester, B. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304, 471–481 (2000).
Champney, W. S. Bacterial ribosomal subunit assembly is an antibiotic target. Curr. Top. Med. Chem. 3, 929–947 (2003).
Katz, L. & Ashley, G. W. Translation and protein synthesis: macrolides. Chem. Rev. 105, 499–528 (2005).
Bryskier, A. Ketolides — telithromycin, an example of a new class of antibacterial agents. Clin. Microbial. Infect. 6, 661–669 (2000).
Farrell, D. J. & Felmingham, D. Activities of telithromycin against 13,874 Streptococcus pneumoniae isolates collected between 1999 and 2003. Antimicrob. Agents Chemother. 48, 1882–1884 (2004).
Schlunzen, F. et al. Structural basis for the antibiotic activity of ketolides and azalides. Structure (Camb.) 11, 329–338 (2003).
Douthwaite, S. & Aagaard, C. Erythromycin binding is reduced in ribosomes with conformational alterations in the 23S rRNA peptidyl transferase loop. 232, 725–731 (1993).
Yonath, A. & Bashan, A. Ribosomal crystallography: initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics. Annu. Rev. Microbiol. 58, 233–251 (2004).
Pfister, P. et al. 23S rRNA base pair 2057–2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A to G. Proc. Natl Acad. Sci. USA 102, 5180–5185 (2005).
Awan, A., Brennan, R. J., Regan, A. C. & Barber, J. The conformations of the macrolide antibiotics erythromycin A, azithromycin and clarithromycin in aqueous solution: a H-1 NMR study. J. Chem. Soc. Perkin Trans. I 2, 1645–1652 (2000).
Zalacain, M. & Cundliffe, E. Cloning of tlrD, a fourth resistance gene, from the tylosin producer, Streptomyces fradiae. Gene 97, 137–142 (1991).
Zalacain, M. & Cundliffe, E. Methylation of 23S rRNA caused by tlrA (ermSF), a tylosin resistance determinant from Streptomyces fradiae. J. Bacteriol. 171, 4254–4260 (1989).
Liu, M. & Douthwaite, S. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc. Natl Acad. Sci. USA 99, 14658–14663 (2002).
Novotny, G. W., Jakobsen, L., Andersen, N. M., Poehlsgaard, J. & Douthwaite, S. Ketolide antimicrobial activity persists after disruption of interactions with domain II of 23S rRNA. Antimicrob. Agents Chemother. 48, 3677–3683 (2004).
Xiong, L., Shah, S., Mauvais, P. & Mankin, A. S. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31, 633–639 (1999).
Hansen, L. H., Mauvais, P. & Douthwaite, S. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31, 623–631 (1999).
Garza-Ramos, G., Xiong, L., Zhong, P. & Mankin, A. Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA. J. Bacteriol. 183, 6898–6907 (2001).
Xiong, L., Korkhin, Y. & Mankin, A. S. Binding site of the bridged macrolides in the Escherichia coli ribosome. Antimicrob. Agents Chemother. 49, 281–288 (2005).
Berisio, R. et al. Structural insight into the antibiotic action of telithromycin against resistant mutants. J. Bacteriol. 185, 4276–4279 (2003).
Vester, B. & Douthwaite, S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45, 1–12 (2001).
Pfister, P. et al. The structural basis of macrolide-ribosome binding assessed using mutagenesis of 23S rRNA positions 2058 and 2059. J. Mol. Biol. 342, 1569–1581 (2004).
Wittmann, H. G. et al. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol. Gen. Genet. 127, 175–189 (1973).
Pardo, D. & Rosset, R. Properties of ribosomes from erythromycin resistant mutants of Escherichia coli. Mol. Gen. Genet. 156, 267–271 (1977).
Chittum, H. S. & Champney, W. S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J. Bacteriol. 176, 6192–6198 (1994).
Tait-Kamradt, A. et al. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44, 2118–2125 (2000).
Farrell, D. J., Morrissey, I., Bakker, S., Buckridge, S. & Felmingham, D. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 48, 3169–3171 (2004).
Gregory, S. T. & Dahlberg, A. E. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J. Mol. Biol. 289, 827–834 (1999).
Davydova, N., Streltsov, V., Wilce, M., Liljas, A. & Garber, M. L22 ribosomal protein and effect of its mutation on ribosome resistance to erythromycin. J. Mol. Biol. 322, 635–644 (2002).
Skinner, R., Cundliffe, E. & Schmidt, F. J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem. 258, 12702–12706 (1983).
Roberts, M. C. Resistance to macrolide, lincosamide, streptogramin, ketolide, and oxazolidinone antibiotics. Mol. Biotechnol. 28, 47–62 (2004).
Liu, M. & Douthwaite, S. Activity of the ketolide telithromycin is refractory to Erm monomethylation of bacterial rRNA. Antimicrob. Agents Chemother. 46, 1629–1633 (2002).
Sutcliffe, J. A. & Leclercq, R. in Macrolide Antibiotics (eds Schönfeld, W. & Kirst, H. A.) 281–317 (Birkhäuser, Berlin, 2002).
Douthwaite, S., Crain, P. F., Liu, M. & Poehlsgaard, J. The tylosin-resistance methyltransferase RlmAII (TlrB) modifies the N-1 position of 23S rRNA nucleotide G748. J. Mol. Biol. 337, 1073–1077 (2004).
Abbanat, D. et al. In vitro activities of novel 2-fluoro-naphthyridine-containing ketolides. Antimicrob. Agents Chemother. 49, 309–315 (2005).
Poehlsgaard, J. & Douthwaite, S. The macrolide binding site on the bacterial ribosome. Curr. Drug Targets Infect. Disord. 2, 67–78 (2002).
Brodersen, D. E. et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103, 1143–1154 (2000).
Vicens, Q. & Westhof, E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem. Biol. 9, 747–755 (2002).
Han, Q. et al. Molecular recognition by glycoside pseudo base pairs and triples in an apramycin–RNA complex. Angew. Chem. (Int. Ed. Engl.) 44, 2694–2700 (2005).
Pioletti, M. et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20, 1829–1839 (2001).
Sutcliffe, J. Rib-X Pharmaceuticals. Antibiotics Conference, Tartu, Estonia, June 2005.
Acknowledgements
Support from the Danish Research Agency and the Nucleic Acid Center of the Danish Grundforskningsfond are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Related links
Glossary
- HAIRPIN
-
A structural element that makes a 180° turn and doubles back on itself; in proteins, normally formed from a β-strand.
- POLYKETIDES
-
Secondary metabolites that are biosynthesized in a stepwise manner from simple 2-, 3- and 4-carbon building blocks; can have antimicrobial, antifungal, antiparasitic, antitumour or agrochemical properties.
- MACROLACTONE RING
-
The cyclic ester ring at the core of macrolide antibiotics.
- MACROLIDE 5-AMINO SUGAR
-
The nitrogen-containing sugar directly attached to the 5-carbon of the macrolactone ring (desosamine in erythromycin and mycaminose in tylosin).
- ELECTROSTATIC INTERACTIONS
-
Interactions between charged molecules or atoms.
- HYDROPHOBIC INTERACTIONS
-
Interactions that rely on the tendency of non-polar groups to aggregate to avoid contact with a polar solvent.
Rights and permissions
About this article
Cite this article
Poehlsgaard, J., Douthwaite, S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol 3, 870–881 (2005). https://doi.org/10.1038/nrmicro1265
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1265
This article is cited by
-
Antimicrobial activity of metal-based nanoparticles: a mini-review
BioMetals (2024)
-
Molecular mechanisms of antibiotic resistance revisited
Nature Reviews Microbiology (2023)
-
16-membered ring macrolides and erythromycin induce ermB expression by different mechanisms
BMC Microbiology (2022)
-
16S rRNA Methyltransferases as Novel Drug Targets Against Tuberculosis
The Protein Journal (2022)
-
Activation of the SigE-SigB signaling pathway by inhibition of the respiratory electron transport chain and its effect on rifampicin resistance in Mycobacterium smegmatis
Journal of Microbiology (2022)