Key Points
-
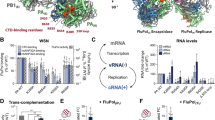
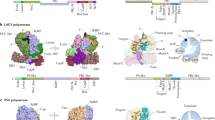
High-resolution structures have recently revealed the subunit and domain organization of the influenza A, influenza B and influenza C virus RNA polymerases. The polymerase basic 1 (PB1) subunit together with the C-terminal domain of polymerase acidic (PA) and the N-terminal one-third of PB2 form the core of the polymerase, which is decorated by several flexible domains, including the PB2 cap-binding and PA endonuclease domains.
-
Crystal structures obtained in the absence and presence of viral genomic RNA have captured the influenza virus RNA polymerase in transcriptionally inactive and transcription pre-initiation states, showing a different arrangement of the peripheral PB2 cap-binding and PA endonuclease domains and suggesting a mechanism for the activation of the cap-snatching function of the polymerase.
-
Structural and biochemical analyses have identified the binding sites of the 5′ and 3′ termini of viral RNAs, the template entry and exit channels, the substrate entry and product exit channels, the polymerase active site and a priming loop.
-
Based on the high-resolution structural information about the influenza virus RNA polymerases, the models for viral transcription and genome replication have been revised.
-
Viral and host factors participate in the regulation of viral transcription and genome replication. However, in many cases, the underlying molecular mechanisms remain unknown.
-
Understanding how the influenza virus RNA synthesis machine works at the molecular level and knowledge of its interaction with viral and host factors might lead to the identification of targets that could be exploited for the development of antivirals to prevent or treat influenza virus infections.
Abstract
The genomes of influenza viruses consist of multiple segments of single-stranded negative-sense RNA. Each of these segments is bound by the heterotrimeric viral RNA-dependent RNA polymerase and multiple copies of nucleoprotein, which form viral ribonucleoprotein (vRNP) complexes. It is in the context of these vRNPs that the viral RNA polymerase carries out transcription of viral genes and replication of the viral RNA genome. In this Review, we discuss our current knowledge of the structure of the influenza virus RNA polymerase, and insights that have been gained into the molecular mechanisms of viral transcription and replication, and their regulation by viral and host factors. Furthermore, we discuss how advances in our understanding of the structure and function of polymerases could help in identifying new antiviral targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Krammer, F. & Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 14, 167–182 (2015).
Watanabe, T. & Kawaoka, Y. Influenza virus–host interactomes as a basis for antiviral drug development. Curr. Opin. Virol. 14, 71–78 (2015).
Krug, R. & Fodor, E. in Textbook of Influenza (eds Webster, R., Monto, A., Braciale, T. & Lamb, R.) 57–66 (Wiley-Blackwell, 2013).
Fodor, E. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta Virol. 57, 113–122 (2013).
Resa-Infante, P., Jorba, N., Coloma, R. & Ortin, J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8, 207–215 (2011).
Arranz, R. et al. The structure of native influenza virion ribonucleoproteins. Science 338, 1634–1637 (2012). This paper describes the 3D structure of native RNPs that are derived from influenza virions.
Moeller, A., Kirchdoerfer, R. N., Potter, C. S., Carragher, B. & Wilson, I. A. Organization of the influenza virus replication machinery. Science 338, 1631–1634 (2012). This work determines the 3D structure of native RNPs that are derived from cells that express influenza virus RNP complex components.
Hutchinson, E. C. & Fodor, E. Transport of the influenza virus genome from nucleus to nucleus. Viruses 5, 2424–2446 (2013).
Plotch, S. J., Bouloy, M., Ulmanen, I. & Krug, R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858 (1981).
Dias, A. et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458, 914–918 (2009).
Guilligay, D. et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15, 500–506 (2008).
Yuan, P. et al. Crystal structure of an avian influenza polymerase PAN reveals an endonuclease active site. Nature 458, 909–913 (2009).
Swale, C. et al. Structural characterization of recombinant IAV polymerase reveals a stable complex between viral PA-PB1 heterodimer and host RanBP5. Sci. Rep. 6, 24727 (2016).
Morin, B., Kranzusch, P. J., Rahmeh, A. A. & Whelan, S. P. J. The polymerase of negative-stranded RNA viruses. Curr. Opin. Virol. 3, 103–110 (2013).
Gerlach, P., Malet, H., Cusack, S. & Reguera, J. Structural insights into bunyavirus replication and its regulation by the vRNA promoter. Cell 161, 1267–1279 (2015).
Hengrung, N. et al. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 527, 114–117 (2015). This report presents the crystal structure of the RNA-free influenza C virus polymerase, revealing a configuration of the PB2 C-terminal domains that is radically different from that in the influenza A and influenza B virus polymerase structures that were obtained in the presence of vRNA. Small-angle X-ray scattering (SAXS) studies show that the polymerase can take various conformations.
Liang, B. et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327 (2015).
Pflug, A., Guilligay, D., Reich, S. & Cusack, S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516, 355–360 (2014). This analysis obtains the crystal structure of a bat influenza A virus polymerase, demonstrating the architecture of the heterotrimeric complex and the structural basis for binding the vRNA promoter, which comprises the conserved 5′ and 3′ termini of the vRNA.
Reich, S. et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516, 361–366 (2014). This investigation determines the crystal structure of the influenza B virus polymerase and shows the rotation of the PB2 cap-binding domain that guides the 3′ end of the capped RNA primer into the polymerase active site.
He, X. et al. Crystal structure of the polymerase PAC–PB1N complex from an avian influenza H5N1 virus. Nature 454, 1123–1126 (2008).
Obayashi, E. et al. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454, 1127–1131 (2008).
Sugiyama, K. et al. Structural insight into the essential PB1–PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28, 1803–1811 (2009).
Tarendeau, F. et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 14, 229–233 (2007).
Tarendeau, F. et al. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 4, e1000136 (2008).
Chang, S. et al. Cryo-EM structure of influenza virus RNA polymerase complex at 4.3 Å resolution. Mol. Cell 57, 925–935 (2015).
te Velthuis, A. J. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 71, 4403–4420 (2014).
Subbarao, E. K., London, W. & Murphy, B. R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67, 1761–1764 (1993).
Thierry, E. et al. Influenza polymerase can adopt an alternative configuration involving a radical repacking of PB2 domains. Mol. Cell 61, 125–137 (2016). This paper presents structures of the influenza B virus polymerase, showing an arrangement of the PB2 C-terminal domains similar to that observed for the influenza C virus polymerase. Solution studies show the polymerase can take up various conformations.
Hsu, M. T., Parvin, J. D., Gupta, S., Krystal, M. & Palese, P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl Acad. Sci. USA 84, 8140–8144 (1987).
Cianci, C., Tiley, L. & Krystal, M. Differential activation of the influenza virus polymerase via template RNA binding. J. Virol. 69, 3995–3999 (1995).
Fodor, E., Pritlove, D. C. & Brownlee, G. G. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68, 4092–4096 (1994).
Fodor, E., Pritlove, D. C. & Brownlee, G. G. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 69, 4012–4019 (1995).
Tiley, L. S., Hagen, M., Matthews, J. T. & Krystal, M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68, 5108–5116 (1994).
Rao, P., Yuan, W. & Krug, R. M. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 22, 1188–1198 (2003).
Tomescu, A. I., Robb, N. C., Hengrung, N., Fodor, E. & Kapanidis, A. N. Single-molecule FRET reveals a corkscrew RNA structure for the polymerase-bound influenza virus promoter. Proc. Natl Acad. Sci. USA 111, E3335–E3342 (2014).
Fodor, E., Seong, B. L. & Brownlee, G. G. Photochemical cross-linking of influenza-a polymerase to its virion RNA promoter defines a polymerase binding-site at residue-9 to residue-12 of the promoter. J. General Virol. 74, 1327–1333 (1993).
Flick, R., Neumann, G., Hoffmann, E., Neumeier, E. & Hobom, G. Promoter elements in the influenza vRNA terminal structure. RNA 2, 1046–1057 (1996).
Pritlove, D. C., Poon, L. L., Devenish, L. J., Leahy, M. B. & Brownlee, G. G. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J. Virol. 73, 2109–2114 (1999).
Pritlove, D. C., Fodor, E., Seong, B. L. & Brownlee, G. G. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J. Gen. Virol. 76, 2205–2213 (1995).
Crow, M., Deng, T., Addley, M. & Brownlee, G. G. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 78, 6263–6270 (2004).
Appleby, T. C. et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347, 771–775 (2015).
Butcher, S. J., Grimes, J. M., Makeyev, E. V., Bamford, D. H. & Stuart, D. L. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410, 235–240 (2001).
Tao, Y. Z., Farsetta, D. L., Nibert, M. L. & Harrison, S. C. RNA synthesis in a cage — Structural studies of reovirus polymerase λ3. Cell 111, 733–745 (2002).
te Velthuis, A. J. W., Robb, N. C., Kapanidis, A. N. & Fodor, E. The role of the priming loop in influenza A virus RNA synthesis. Nat. Microbiol. 1, 16029 (2016). This research finds that the priming loop of the influenza A virus polymerase is required for terminal de novo replication initiation on the vRNA template, but is not required for internal de novo replication initiation on the cRNA template and for capped RNA primer-dependent transcription initiation.
Gu, W. et al. Influenza A virus preferentially snatches noncoding RNA caps. RNA 21, 2067–2075 (2015).
Koppstein, D., Ashour, J. & Bartel, D. P. Sequencing the cap-snatching repertoire of H1N1 influenza provides insight into the mechanism of viral transcription initiation. Nucleic Acids Res. 43, 5052–5064 (2015).
Sikora, D., Rocheleau, L., Brown, E. G. & Pelchat, M. Deep sequencing reveals the eight facets of the influenza A/HongKong/1/1968 (H3N2) virus cap-snatching process. Sci. Rep. 4, 6181 (2014).
Beaton, A. R. & Krug, R. M. Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res. 9, 4423–4436 (1981).
Shaw, M. W. & Lamb, R. A. A specific sub-set of host-cell mRNAs prime influenza virus mRNA synthesis. Virus Res. 1, 455–467 (1984).
Poon, L. L., Pritlove, D. C., Fodor, E. & Brownlee, G. G. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73, 3473–3476 (1999).
Poon, L. L., Fodor, E. & Brownlee, G. G. Polyuridylated mRNA synthesized by a recombinant influenza virus is defective in nuclear export. J. Virol. 74, 418–427 (2000).
Braam, J., Ulmanen, I. & Krug, R. M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34, 609–618 (1983).
Bier, K., York, A. & Fodor, E. Cellular cap-binding proteins associate with influenza virus mRNAs. J. Gen. Virol. 92, 1627–1634 (2011).
York, A. & Fodor, E. Biogenesis, assembly, and export of viral messenger ribonucleoproteins in the influenza A virus infected cell. RNA Biol. 10, 1274–1282 (2013).
Vreede, F. T. & Brownlee, G. G. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J. Virol. 81, 2196–2204 (2007).
York, A., Hengrung, N., Vreede, F. T., Huiskonen, J. T. & Fodor, E. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc. Natl Acad. Sci. USA 110, E4238–E4245 (2013). This article reports the isolation and subsequent structural and functional characterization of the influenza A virus cRNP replicative intermediate. The authors propose that the cRNP-associated polymerase requires a trans -activating polymerase to be able to carry out vRNA synthesis.
Deng, T., Vreede, F. T. & Brownlee, G. G. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 80, 2337–2348 (2006).
Chan, W. H. et al. Functional analysis of the influenza virus H5N1 nucleoprotein tail loop reveals amino acids that are crucial for oligomerization and ribonucleoprotein activities. J. Virol. 84, 7337–7345 (2010).
Ng, A. K. et al. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 22, 3638–3647 (2008).
Ye, Q., Krug, R. M. & Tao, Y. J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444, 1078–1082 (2006).
Turrell, L., Lyall, J. W., Tiley, L. S., Fodor, E. & Vreede, F. T. The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nat. Commun. 4, 1591 (2013).
Boulo, S. et al. Human importin-α and RNA do not compete for binding to influenza A virus nucleoprotein. Virology 409, 84–90 (2011).
Chenavas, S. et al. Monomeric nucleoprotein of influenza A virus. PLoS Pathog. 9, e1003275 (2013).
Mondal, A., Potts, G. K., Dawson, A. R., Coon, J. J. & Mehle, A. Phosphorylation at the homotypic interface regulates nucleoprotein oligomerization and assembly of the influenza virus replication machinery. PLoS Pathog. 11, e1004826 (2015).
Turrell, L., Hutchinson, E. C., Vreede, F. T. & Fodor, E. Regulation of influenza A virus nucleoprotein oligomerization by phosphorylation. J. Virol. 89, 1452–1455 (2015).
Jorba, N., Coloma, R. & Ortin, J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 5, e1000462 (2009). This report describes a model for influenza virus transcription and genome replication. It proposes that transcription is carried out by the resident polymerase bound to the vRNA termini in the vRNP, whereas the cRNA-to-vRNA step of genome replication is carried out by a trans -acting polymerase that is not part of the cRNP.
Beaton, A. R. & Krug, R. M. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc. Natl Acad. Sci. USA 83, 6282–6286 (1986).
Shapiro, G. I. & Krug, R. M. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J. Virol. 62, 2285–2290 (1988).
Vreede, F. T., Jung, T. E. & Brownlee, G. G. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78, 9568–9572 (2004).
Resa-Infante, P., Recuero-Checa, M. A., Zamarreno, N., Llorca, O. & Ortin, J. Structural and functional characterization of an influenza virus RNA polymerase-genomic RNA complex. J. Virol. 84, 10477–10487 (2010).
Kawaguchi, A., Momose, F. & Nagata, K. Replication-coupled and host factor-mediated encapsidation of the influenza virus genome by viral nucleoprotein. J. Virol. 85, 6197–6204 (2011).
Newcomb, L. L. et al. Interaction of the influenza a virus nucleocapsid protein with the viral RNA polymerase potentiates unprimed viral RNA replication. J. Virol. 83, 29–36 (2009).
Hutchinson, E. C. et al. Conserved and host-specific features of influenza virion architecture. Nat. Commun. 5, 4816 (2014).
Min, J. Y., Li, S., Sen, G. C. & Krug, R. M. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363, 236–243 (2007).
Robb, N. C. et al. The influenza A virus NS1 protein interacts with the nucleoprotein of viral ribonucleoprotein complexes. J. Virol. 85, 5228–5231 (2011).
Marion, R. M., Zurcher, T., de la Luna, S. & Ortin, J. Influenza virus NS1 protein interacts with viral transcription–replication complexes in vivo. J. Gen. Virol. 78, 2447–2451 (1997).
Falcon, A. M. et al. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78, 3880–3888 (2004).
Garaigorta, U. & Ortin, J. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 35, 4573–4582 (2007).
Robb, N. C. & Fodor, E. The accumulation of influenza A virus segment 7 spliced mRNAs is regulated by the NS1 protein. J. Gen. Virol. 93, 113–118 (2012).
Paterson, D. & Fodor, E. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 8, e1003019 (2012).
Robb, N. C., Smith, M., Vreede, F. T. & Fodor, E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90, 1398–1407 (2009).
Manz, B., Brunotte, L., Reuther, P. & Schwemmle, M. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat. Commun. 3, 802 (2012).
Perez, J. T. et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl Acad. Sci. USA 107, 11525–11530 (2010).
Umbach, J. L., Yen, H. L., Poon, L. L. & Cullen, B. R. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio 1, e00204–10 (2010).
Perez, J. T. et al. A small-RNA enhancer of viral polymerase activity. J. Virol. 86, 13475–13485 (2012).
Brass, A. L. et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 (2009).
Hao, L. et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454, 890–893 (2008).
Karlas, A. et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822 (2010).
Shapira, S. D. et al. A physical and regulatory map of host–influenza interactions reveals pathways in H1N1 infection. Cell 139, 1255–1267 (2009).
Naito, T. et al. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc. Natl Acad. Sci. USA 104, 18235–18240 (2007).
Sui, B. et al. The use of Random Homozygous Gene Perturbation to identify novel host-oriented targets for influenza. Virology 387, 473–481 (2009).
Tripathi, S. et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe 18, 723–735 (2015).
Watanabe, T. et al. Influenza virus–host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16, 795–805 (2014).
Konig, R. et al. Human host factors required for influenza virus replication. Nature 463, 813–817 (2010).
Jorba, N. et al. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8, 2077–2088 (2008).
York, A., Hutchinson, E. C. & Fodor, E. Interactome analysis of the influenza A virus transcription/replication machinery identifies protein phosphatase 6 as a cellular factor required for efficient virus replication. J. Virol. 88, 13284–13299 (2014).
Mayer, D. et al. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6, 672–682 (2007).
Bradel-Tretheway, B. G. et al. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. J. Virol. 85, 8569–8581 (2011).
Bortz, E. et al. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio 2, e00151–11 (2011).
Cao, M. et al. DnaJA1/Hsp40 is co-opted by influenza A virus to enhance its viral RNA polymerase activity. J. Virol. 88, 14078–14089 (2014).
Deng, T. et al. Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 80, 11911–11919 (2006).
Fislova, T., Thomas, B., Graef, K. M. & Fodor, E. Association of the influenza virus RNA polymerase subunit PB2 with the host chaperonin CCT. J. Virol. 84, 8691–8699 (2010).
Hutchinson, E. C., Orr, O. E., Man Liu, S., Engelhardt, O. G. & Fodor, E. Characterization of the interaction between the influenza A virus polymerase subunit PB1 and the host nuclear import factor Ran-binding protein 5. J. Gen. Virol. 92, 1859–1869 (2011).
Chase, G. P. et al. Influenza virus ribonucleoprotein complexes gain preferential access to cellular export machinery through chromatin targeting. PLoS Pathog. 7, e1002187 (2011).
Jackson, D. A., Caton, A. J., McCready, S. J. & Cook, P. R. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature 296, 366–368 (1982).
Lopez-Turiso, J. A., Martinez, C., Tanaka, T. & Ortin, J. The synthesis of influenza virus negative-strand RNA takes place in insoluble complexes present in the nuclear matrix fraction. Virus Res. 16, 325–337 (1990).
Takizawa, N., Watanabe, K., Nouno, K., Kobayashi, N. & Nagata, K. Association of functional influenza viral proteins and RNAs with nuclear chromatin and sub-chromatin structure. Microbes Infect. 8, 823–833 (2006).
Alfonso, R. et al. CHD6 chromatin remodeler is a negative modulator of influenza virus replication that relocates to inactive chromatin upon infection. Cell. Microbiol. 13, 1894–1906 (2011).
Marcos-Villar, L., Pazo, A. & Nieto, A. Influenza virus and chromatin: role of the CHD1 chromatin remodeler in the virus life cycle. J. Virol. 90, 3694–3707 (2016).
Rodriguez, A., Perez-Gonzalez, A. & Nieto, A. Cellular human CLE/C14orf166 protein interacts with influenza virus polymerase and is required for viral replication. J. Virol. 85, 12062–12066 (2011).
Ver, L. S., Marcos-Villar, L., Landeras-Bueno, S., Nieto, A. & Ortin, J. The cellular factor NXP2/MORC3 is a positive regulator of influenza virus multiplication. J. Virol. 89, 10023–10030 (2015).
Martinez-Alonso, M., Hengrung, N. & Fodor, E. RNA-free and ribonucleoprotein-associated influenza virus polymerases directly bind the serine-5 phosphorylated carboxyl-terminal domain of host RNA polymerase II. J. Virol. 90, 6014–6021 (2016).
Engelhardt, O. G., Smith, M. & Fodor, E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 79, 5812–5818 (2005). This is the first study to demonstrate the interaction between the influenza virus RNA polymerase and the host Pol II transcriptional machinery. The authors propose that cap-snatching is carried out by the viral polymerase bound to the serine-5-phosphorylated form of the CTD of Pol II.
Bentley, D. L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 (2014).
Yamayoshi, S., Watanabe, M., Goto, H. & Kawaoka, Y. Identification of a novel viral protein expressed from the PB2 segment of influenza A virus. J. Virol. 90, 444–456 (2015).
Fournier, G. et al. Recruitment of RED–SMU1 complex by influenza A virus RNA polymerase to control viral mRNA splicing. PLoS Pathog. 10, e1004164 (2014).
Landeras-Bueno, S., Jorba, N., Perez-Cidoncha, M. & Ortin, J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 7, e1002397 (2011).
Shih, S. R. & Krug, R. M. Novel exploitation of a nuclear function by influenza virus: the cellular SF2/ASF splicing factor controls the amount of the essential viral M2 ion channel protein in infected cells. EMBO J. 15, 5415–5427 (1996).
Tsai, P. L. et al. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathog. 9, e1003460 (2013).
Kawaguchi, A. & Nagata, K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 26, 4566–4575 (2007).
Sugiyama, K., Kawaguchi, A., Okuwaki, M. & Nagata, K. pp32 and APRIL are host cell-derived regulators of influenza virus RNA synthesis from cRNA. eLife 4, e08939 (2015). This analysis identifies ANP32A and ANP32B as host factors that are required for the replication of the influenza A virus cRNA replicative intermediate into vRNA.
Long, J. S. et al. Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature 529, 101–104 (2016). This work finds that ANP32A is the host factor that underlies the influenza A virus polymerase host restriction mediated by residue 627 of the PB2 subunit.
Zhou, Z. et al. Fragile X mental retardation protein stimulates ribonucleoprotein assembly of influenza A virus. Nat. Commun. 5, 3259 (2014).
Resa-Infante, P. et al. The host-dependent interaction of α-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS ONE 3, e3904 (2008).
Nemeroff, M. E., Barabino, S. M., Li, Y., Keller, W. & Krug, R. M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1, 991–1000 (1998).
Vreede, F. T. & Fodor, E. The role of the influenza virus RNA polymerase in host shut-off. Virulence 1, 436–439 (2010).
Vreede, F. T., Chan, A. Y., Sharps, J. & Fodor, E. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 396, 125–134 (2010).
Llompart, C. M., Nieto, A. & Rodriguez-Frandsen, A. Specific residues of PB2 and PA influenza virus polymerase subunits confer the ability for RNA polymerase II degradation and virus pathogenicity in mice. J. Virol. 88, 3455–3463 (2014).
Jorba, N., Area, E. & Ortin, J. Oligomerization of the influenza virus polymerase complex in vivo. J. Gen. Virol. 89, 520–524 (2008).
Vasin, A. V. et al. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res. 185, 53–63 (2014).
Monod, A. et al. Learning from structure-based drug design and new antivirals targeting the ribonucleoprotein complex for the treatment of influenza. Expert Opin. Drug Discov. 10, 345–371 (2015).
Byrn, R. A. et al. Preclinical activity of VX-787, a first-in-class, orally bioavailable inhibitor of the influenza virus polymerase PB2 subunit. Antimicrob. Agents Chemother. 59, 1569–1582 (2015).
Clark, M. P. et al. Discovery of a novel, first-in-class, orally bioavailable azaindole inhibitor (VX-787) of influenza PB2. J. Med. Chem. 57, 6668–6678 (2014).
DuBois, R. M. et al. Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 8, e1002830 (2012).
Kowalinski, E. et al. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 polymerase. PLoS Pathog. 8, e1002831 (2012).
Pautus, S. et al. New 7-methylguanine derivatives targeting the influenza polymerase PB2 cap-binding domain. J. Med. Chem. 56, 8915–8930 (2013).
Sagong, H. Y. et al. Phenyl substituted 4-hydroxypyridazin-3(2H)-ones and 5-hydroxypyrimidin-4(3H)-ones: inhibitors of influenza A endonuclease. J. Med. Chem. 57, 8086–8098 (2014).
Yuan, S. et al. A novel small-molecule inhibitor of influenza A virus acts by suppressing PA endonuclease activity of the viral polymerase. Sci. Rep. 6, 22880 (2016).
Sangawa, H. et al. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob. Agents Chemother. 57, 5202–5208 (2013).
Sleeman, K. et al. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 A(H1N1) viruses. Antimicrob. Agents Chemother. 54, 2517–2524 (2010).
Tisdale, M. et al. Inhibition of influenza A and B viruses by 2′-deoxy-2′-fluororibosides. Antiviral Chem. Chemother. 4, 281–287 (1993).
Ghanem, A. et al. Peptide-mediated interference with influenza A virus polymerase. J. Virol. 81, 7801–7804 (2007).
Massari, S. et al. A broad anti-influenza hybrid small molecule that potently disrupts the interaction of polymerase acidic protein–basic protein 1 (PA-PB1) subunits. J. Med. Chem. 58, 3830–3842 (2015).
Wunderlich, K. et al. Identification of high-affinity PB1-derived peptides with enhanced affinity to the PA protein of influenza A virus polymerase. Antimicrob. Agents Chemother. 55, 696–702 (2011).
Yuan, S. et al. Identification of a small-molecule inhibitor of influenza virus via disrupting the subunits interaction of the viral polymerase. Antiviral Res. 125, 34–42 (2016).
Manz, B. et al. Disruption of the viral polymerase complex assembly as a novel approach to attenuate influenza A virus. J. Biol. Chem. 286, 8414–8424 (2011).
Muratore, G. et al. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc. Natl Acad. Sci. USA 109, 6247–6252 (2012).
Tintori, C. et al. High-throughput docking for the identification of new influenza A virus polymerase inhibitors targeting the PA–PB1 protein–protein interaction. Bioorg. Med. Chem. Lett. 24, 280–282 (2014).
Li, C. et al. A peptide derived from the C-terminus of PB1 inhibits influenza virus replication by interfering with viral polymerase assembly. FEBS J. 280, 1139–1149 (2013).
Acknowledgements
The authors are grateful to F. Vreede for his comments on the manuscript. This work was supported by the UK Medical Research Council grant MR/K000241/1 (to E.F.), the Wellcome Trust grant 098721/Z/12/Z (to A.T.), the Netherlands Organization for Scientific Research (NWO) grant 825.11.029 (to A.T.) and a Kemp postdoctoral fellowship from Lincoln College, University of Oxford, UK (to A.T.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Supplementary information
Supplementary information S1 (movie)
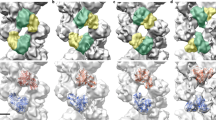
Conformational flexibility of the influenza virus RNA polymerase. Morph between the structure of the RNA-free influenza C virus polymerase (PDB: 5D98) and that of vRNA promoter-bound influenza A virus polymerase (PDB: 4WSB). PB1 and PA are coloured orange and blue, respectively. PB2 is coloured according to its domain structure with the N terminal one third shown in green, the cap-binding domain in dark yellow, the mid-link in light yellow, the 627 domain in red and the C terminal NLS in brown. Promoter RNA is shown in red. Modified from Hengrung et al. (MOV 14521 kb)
Glossary
- Antigenic variation
-
Changes in the antigenicity of a protein of interest (here, the haemagglutinin and neuraminidase proteins) owing to antigenic drift or shift.
- Haemagglutinin
-
(HA). A type I integral membrane glycoprotein of influenza viruses that binds to cell surface receptors in the host and facilitates fusion between the viral envelope and endosomal membrane. It is the main antigen that is targeted by the humoral immune response of the host.
- Neuraminidase
-
(NA). A type II integral membrane glycoprotein of influenza viruses that facilitates viral release from cells by removing sialic acid from sialyloligosaccharides on the cell and viral surface.
- Cap-snatching
-
A process in which a cellular capped RNA is cleaved a few nucleotides downstream of the 5′-cap by an endonuclease activity that is encompassed within a viral RNA-dependent RNA polymerase.
- Host range determinant
-
A characteristic of a pathogen that enables it to replicate in a particular host.
- Panhandle structure
-
A double-stranded RNA structure that is formed by the conserved 5′ and 3′ RNA termini of negative-sense viral RNA genomes.
- Apo form
-
A protein with no ligand bound.
- Helicase
-
An enzyme that catalyses the unwinding and separation of double-stranded DNA or RNA using energy from ATP hydrolysis.
- De novo initiation
-
The initiation step of nucleic acid synthesis that occurs by means of a primer-independent mechanism.
- Pol II pausing
-
(Cellular DNA-dependent RNA polymerase II pausing). A control step in gene transcription, in which Pol II pauses at certain sites and requires specific stimuli and elongation factors to overcome this pausing block and enter productive elongation.
- Pol II large subunit CTD
-
(Cellular DNA-dependent RNA polymerase II large subunit C-terminal domain). An unstructured, evolutionarily conserved domain at the C terminus of the largest Pol II subunit; this domain comprises tandem copies of the consensus heptapeptide YSPTSPS, and phosphorylation of these repeats is crucial for the regulation of Pol II function.
Rights and permissions
About this article
Cite this article
te Velthuis, A., Fodor, E. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol 14, 479–493 (2016). https://doi.org/10.1038/nrmicro.2016.87
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2016.87
This article is cited by
-
Mechanisms and consequences of mRNA destabilization during viral infections
Virology Journal (2024)
-
The ubiquitination landscape of the influenza A virus polymerase
Nature Communications (2023)
-
Phosphorylation of the PA subunit of influenza polymerase at Y393 prevents binding of the 5′-termini of RNA and polymerase function
Scientific Reports (2023)
-
Microbiota-derived acetate enhances host antiviral response via NLRP3
Nature Communications (2023)
-
A review of immune modulators and immunotherapy in infectious diseases
Molecular and Cellular Biochemistry (2023)