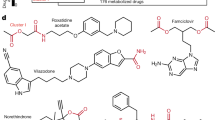
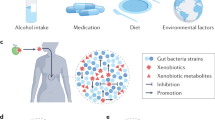
Key Points
-
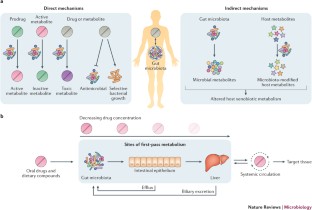
The gut microbiome is a neglected component of the first-pass metabolism of xenobiotics before reaching the general circulation.
-
Direct microbial metabolism of xenobiotics and their metabolites often involves reduction or hydrolysis, but most of the enzymes responsible for these reactions remain unknown.
-
Microbial metabolism influences both efficacy and toxicity, producing bioactive compounds, inactive metabolites and toxins.
-
Relevant host–microbial interactions include the expression of host genes that are involved in drug transport and metabolism, the interference with host enzymatic activity and the modulation of immune responses.
-
The translational implications of these studies include the development of novel co-therapies and the identification of new biomarkers and drugs.
Abstract
Although the importance of human genetic polymorphisms in therapeutic outcomes is well established, the role of our 'second genome' (the microbiome) has been largely overlooked. In this Review, we highlight recent studies that have shed light on the mechanisms that link the human gut microbiome to the efficacy and toxicity of xenobiotics, including drugs, dietary compounds and environmental toxins. Continued progress in this area could enable more precise tools for predicting patient responses and for the development of a new generation of therapeutics based on, or targeted at, the gut microbiome. Indeed, the admirable goal of precision medicine may require us to first understand the microbial pharmacists within.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Faith, J. J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Kuczynski, J. et al. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 13, 47–58 (2012).
Maurice, C. F., Haiser, H. J. & Turnbaugh, P. J. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013). This study is the first to develop methods to define the metabolically active set of gut bacteria and demonstrate that xenobiotics shape the structure and physiology of these bacteria.
Maurice, C. F. & Turnbaugh, P. J. Quantifying the metabolic activities of human-associated microbial communities across multiple ecological scales. FEMS Microbiol. Rev. 37, 830–848 (2013).
O'Hara, A. M. & Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 7, 688–693 (2006).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Thaiss, C. A. et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529 (2014).
Diaz Heijtz, R. et al. Normal gut microbiota modulates brain development and behavior. Proc. Natl Acad. Sci. USA 108, 3047–3052 (2011).
Foster, J. A. & McVey Neufeld, K. A. Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312 (2013).
Hsiao, E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013).
Fierer, N. et al. Forensic identification using skin bacterial communities. Proc. Natl Acad. Sci. USA 107, 6477–6481 (2010).
Franzosa, E. A. et al. Identifying personal microbiomes using metagenomic codes. Proc. Natl Acad. Sci. USA 112, E2930–E2938 (2015).
Liou, A. P. et al. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci. Transl. Med. 5, 178ra141 (2013).
Roopchand, D. E. et al. Dietary polyphenols promote growth of the gut bacterium Akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes 64, 2847–2858 (2015). This study suggests that the beneficial effects of dietary polyphenols may be mediated by the gut microbiome.
Fuller, A. T. Is p-aminobenzenesulphonamide the active agent in protonsil therapy? Lancet 229, 194–198 (1937).
Colebrook, L., Buttle, G. A. H. & O'Meara, R. A. Q. The mode of action of p-aminobenzene sulphonamide and prontosil in hemolytic streptococcal infections. Lancet 228, 1323–1326 (1936).
Radomski, J. L. & Mellinger, T. J. The absorption, fate and excretion in rats of the water-soluble azo dyes, FD&C Red No. 2, FD&C Red No. 4, and FD&C Yellow No. 6. J. Pharmacol. Exp. Ther. 136, 259–266 (1962).
Klotz, U., Maier, K., Fischer, C. & Heinkel, K. Therapeutic efficacy of sulfasalazine and its metabolites in patients with ulcerative colitis and Crohn's disease. N. Engl. J. Med. 303, 1499–1502 (1980).
Plosker, G. L. & Croom, K. F. Sulfasalazine: a review of its use in the management of rheumatoid arthritis. Drugs 65, 1825–1849 (2005).
Rocco, T. P. & Fang, J. C. in Goodman & Gilman's The Pharmacological Basis of Therapeutics (eds Brunton, L. L., Lazo, J. S. & Parker, K. L.) (McGraw-Hill, 2011).
Grundmann, O. The gut microbiome and pre-systemic metabolism: current state and evolving research. J. Drug Metab. Toxicol. 1, 1–7 (2010).
Haiser, H. J. & Turnbaugh, P. J. Developing a metagenomic view of xenobiotic metabolism. Pharmacol. Res. 69, 21–31 (2013).
Li, H. & Jia, W. Cometabolism of microbes and host: implications for drug metabolism and drug-induced toxicity. Clin. Pharmacol. Ther. 94, 574–581 (2013).
Nicholson, J. K., Holmes, E. & Wilson, I. D. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3, 431–438 (2005).
Saad, R., Rizkallah, M. R. & Aziz, R. K. Gut pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 4, 16 (2012).
Tralau, T., Sowada, J. & Luch, A. Insights on the human microbiome and its xenobiotic metabolism: what is known about its effects on human physiology? Expert Opin. Drug Metab. Toxicol. 11, 411–425 (2015).
Pond, S. M. & Tozer, T. N. First-pass elimination. Basic concepts and clinical consequences. Clin. Pharmacokinet. 9, 1–25 (1984).
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host–bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Arkhipova, O. V. & Akimenko, V. K. Unsaturated organic acids as terminal electron acceptors for reductase chains of anaerobic bacteria. Microbiology 76, 725–737 (2005).
Novel, G., Didier-Fichet, M. L. & Stoeber, F. Inducibility of β-glucuronidase in wild-type and hexuronate-negative mutants of Escherichia coli K-12. J. Bacteriol. 120, 89–95 (1974).
Haiser, H. J., Seim, K. L., Balskus, E. P. & Turnbaugh, P. J. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes 5, 233–238 (2014).
de Groot, M. J. Designing better drugs: predicting cytochrome P450 metabolism. Drug Discov. Today 11, 601–606 (2006).
Zanger, U. M. & Schwab, M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141 (2013).
Bachrach, W. H. Sulfasalazine: I. An historical perspective. Am. J. Gastroenterol. 83, 487–496 (1988).
Svartz, N. Sulfasalazine: II. Some notes on the discovery and development of salazopyrin. Am. J. Gastroenterol. 83, 497–503 (1988).
Peppercorn, M. A. & Goldman, P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J. Pharmacol. Exp. Ther. 181, 555–562 (1972).
Chen, H., Wang, R. F. & Cerniglia, C. E. Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr. Purif. 34, 302–310 (2004).
Morrison, J. M., Wright, C. M. & John, G. H. Identification, isolation and characterization of a novel azoreductase from Clostridium perfringens. Anaerobe 18, 229–234 (2012).
Sousa, T. et al. On the colonic bacterial metabolism of azo-bonded prodrugs of 5-aminosalicylic acid. J. Pharm. Sci. 103, 3171–3175 (2014).
Delomenie, C. et al. Identification and functional characterization of arylamine N-acetyltransferases in eubacteria: evidence for highly selective acetylation of 5-aminosalicylic acid. J. Bacteriol. 183, 3417–3427 (2001).
Carmody, R. N. & Turnbaugh, P. J. Host–microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Invest. 124, 4173–4181 (2014).
Wells, P. G. et al. Glucuronidation and the UDP-glucuronosyltransferases in health and disease. Drug Metab. Dispos. 32, 281–290 (2004).
Wiseman, L. R. & Markham, A. Irinotecan. A review of its pharmacological properties and clinical efficacy in the management of advanced colorectal cancer. Drugs 52, 606–623 (1996).
Stein, A., Voigt, W. & Jordan, K. Chemotherapy-induced diarrhea: pathophysiology, frequency and guideline-based management. Ther. Adv. Med. Oncol. 2, 51–63 (2010).
Mani, S., Boelsterli, U. A. & Redinbo, M. R. Understanding and modulating mammalian–microbial communication for improved human health. Annu. Rev. Pharmacol. Toxicol. 54, 559–580 (2014).
Rothenberg, M. L. et al. Phase II trial of irinotecan in patients with progressive or rapidly recurrent colorectal cancer. J. Clin. Oncol. 14, 1128–1135 (1996).
Higuchi, K. et al. Present status and strategy of NSAIDs-induced small bowel injury. J. Gastroenterol. 44, 879–888 (2009).
Saitta, K. S. et al. Bacterial β-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica 44, 28–35 (2014). This study demonstrates that the toxicity associated with NSAIDs can be alleviated by inhibiting bacterial enzyme activity with small-molecule inhibitors.
Beaud, D., Tailliez, P. & Anba-Mondoloni, J. Genetic characterization of the β-glucuronidase enzyme from a human intestinal bacterium, Ruminococcus gnavus. Microbiology 151, 2323–2330 (2005).
Dabek, M., McCrae, S. I., Stevens, V. J., Duncan, S. H. & Louis, P. Distribution of β-glucosidase and β-glucuronidase activity and of β-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 66, 487–495 (2008).
Flores, R. et al. Association of fecal microbial diversity and taxonomy with selected enzymatic functions. PLoS ONE 7, e39745 (2012).
Roy, D. & Ward, P. Rapid detection of Bifidobacterium dentium by enzymatic hydrolysis of β-glucuronide substrates. J. Food Protect. 55, 291–295 (1992).
Russell, W. M. & Klaenhammer, T. R. Identification and cloning of gusA, encoding a new β-glucuronidase from Lactobacillus gasseri ADH. Appl. Environ. Microbiol. 67, 1253–1261 (2001).
Wallace, B. D. et al. Structure and inhibition of microbiome β-glucuronidases essential to the alleviation of cancer drug toxicity. Chem. Biol. 22, 1238–1249 (2015).
Lindenbaum, J. et al. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N. Engl. J. Med. 305, 789–794 (1981).
Matzuk, M. M., Shlomchik, M. & Shaw, L. M. Making digoxin therapeutic drug monitoring more effective. Ther. Drug Monit. 13, 215–219 (1991).
Peters, U., Falk, L. C. & Kalman, S. M. Digoxin metabolism in patients. Arch. Intern. Med. 138, 1074–1076 (1978).
Saha, J. R., Butler, V. P., Neu, H. C. & Lindenbaum, J. Digoxin-inactivating bacteria: identification in human gut flora. Science 220, 325–327 (1983).
Mathan, V. I., Wiederman, J., Dobkin, J. F. & Lindenbaum, J. Geographic differences in digoxin inactivation, a metabolic activity of the human anaerobic gut flora. Gut 30, 971–977 (1989).
Rowland, I. R. Factors affecting metabolic activity of the intestinal microflora. Drug Metab. Rev. 19, 243–261 (1988).
Haiser, H. J. et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298 (2013). This study is the first to show that the bacterial inactivation of drugs can be predicted with a genetic marker and can be prevented using dietary intervention.
Hooper, L. V. et al. Molecular analysis of commensal host–microbial relationships in the intestine. Science 291, 881–884 (2001).
Bjorkholm, B. et al. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS ONE 4, e6958 (2009).
Lundin, A. et al. Gut flora, Toll-like receptors and nuclear receptors: a tripartite communication that tunes innate immunity in large intestine. Cell. Microbiol. 10, 1093–1103 (2008).
Claus, S. P. et al. Colonization-induced host–gut microbial metabolic interaction. mBio 2, e00271-10 (2011).
Selwyn, F. P., Cui, J. Y. & Klaassen, C. D. RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab. Dispos. 43, 1572–1580 (2015).
Claus, S. P. et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4, 219 (2008).
Wikoff, W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. USA 106, 3698–3703 (2009). This study demonstrates that the microbiome has a substantial role in the signature and abundance of circulating metabolites in mammalian blood.
Hodgman, M. J. & Garrard, A. R. A review of acetaminophen poisoning. Crit. Care Clin. 28, 499–516 (2012).
Court, M. H. et al. Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms. J. Pharmacol. Exp. Ther. 299, 998–1006 (2001).
Harrill, A. H. et al. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 19, 1507–1515 (2009).
Clayton, T. A., Baker, D., Lindon, J. C., Everett, J. R. & Nicholson, J. K. Pharmacometabonomic identification of a significant host–microbiome metabolic interaction affecting human drug metabolism. Proc. Natl Acad. Sci. USA 106, 14728–14733 (2009). This study is the first to demonstrate the potential of using levels of microbial metabolites as predictive biomarkers for drug metabolism.
Bone, E., Tamm, A. & Hill, M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am. J. Clin. Nutr. 29, 1448–1454 (1976).
Selmer, T. & Andrei, P. I. p-hydroxyphenylacetate decarboxylase from Clostridium difficile. A novel glycyl radical enzyme catalysing the formation of p-cresol. Eur. J. Biochem. 268, 1363–1372 (2001).
Gamage, N. et al. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 90, 5–22 (2006).
Mangravite, L. M., Thorn, C. F. & Krauss, R. M. Clinical implications of pharmacogenomics of statin treatment. Pharmacogenom. J. 6, 360–374 (2006).
Kaddurah-Daouk, R. et al. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS ONE 6, e25482 (2011).
Mitchell, J. B. et al. A low molecular weight antioxidant decreases weight and lowers tumor incidence. Free Radic. Biol. Med. 34, 93–102 (2003).
Li, F. et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 4, 2384 (2013).
Jiang, J. et al. Diversity of bile salt hydrolase activities in different lactobacilli toward human bile salts. Ann. Microbiol. 60, 81–88 (2010).
de Wit, N. J. et al. The role of the small intestine in the development of dietary fat-induced obesity and insulin resistance in C57BL/6J mice. BMC Med. Genom. 1, 14 (2008).
Sayin, S. I. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-β-muricholic acid, a naturally occurring FXR antagonist. Cell. Metab. 17, 225–235 (2013).
Tsuda, T. Regulation of adipocyte function by anthocyanins; possibility of preventing the metabolic syndrome. J. Agr. Food Chem. 56, 642–646 (2008).
Quesada, H. et al. Grape seed proanthocyanidins correct dyslipidemia associated with a high-fat diet in rats and repress genes controlling lipogenesis and VLDL assembling in liver. Int. J. Obes. (Lond.) 33, 1007–1012 (2009).
Tsuda, T., Horio, F., Uchida, K., Aoki, H. & Osawa, T. Dietary cyanidin 3-O-β-d-glucoside-rich purple corn color prevents obesity and ameliorates hyperglycemia in mice. J. Nutr. 133, 2125–2130 (2003).
Baiges, I., Palmfeldt, J., Blade, C., Gregersen, N. & Arola, L. Lipogenesis is decreased by grape seed proanthocyanidins according to liver proteomics of rats fed a high fat diet. Mol. Cell. Proteom. 9, 1499–1513 (2010).
Cefalu, W. T. et al. Botanicals and the metabolic syndrome. Am. J. Clin. Nutr. 87, 481S–487S (2008).
Felgines, C. et al. Radiolabelled cyanidin 3-O-glucoside is poorly absorbed in the mouse. Br. J. Nutr. 103, 1738–1745 (2010).
Abia, R. & Fry, S. C. Degradation and metabolism of 14C-labelled proanthocyanidins from carob (Ceratonia siliqua) pods in the gastrointestinal tract of the rat. J. Sci. Food Agr. 81, 1156–1165 (2001).
Anhe, F. F. et al. A polyphenol-rich cranberry extract protects from diet-induced obesity, insulin resistance and intestinal inflammation in association with increased Akkermansia spp. population in the gut microbiota of mice. Gut 64, 872–883 (2015).
Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl Acad. Sci. USA 110, 9066–9071 (2013).
Santacruz, A. et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 104, 83–92 (2010).
Zhang, H. et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl Acad. Sci. USA 106, 2365–2370 (2009).
Everard, A. et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60, 2775–2786 (2011).
Shin, N. R. et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735 (2014). This study suggests that the anti-diabetic drug metformin may act, in part, by altering the gut microbiome.
Axling, U. et al. Green tea powder and Lactobacillus plantarum affect gut microbiota, lipid metabolism and inflammation in high-fat fed C57BL/6J mice. Nutr. Metab. (Lond.) 9, 105 (2012).
Kemperman, R. A. et al. Impact of polyphenols from black tea and red wine/grape juice on a gut model microbiome. Food Res. Int. 53, 659–669 (2013).
Haslam, E. Natural polyphenols (vegetable tannins) as drugs: possible modes of action. J. Nat. Prod. 59, 205–215 (1996).
Nunez-Sanchez, M. A. et al. Targeted metabolic profiling of pomegranate polyphenols and urolithins in plasma, urine and colon tissues from colorectal cancer patients. Mol. Nutr. Food. Res. 58, 1199–1211 (2014).
Garcia-Munoz, C. & Vaillant, F. Metabolic fate of ellagitannins: implications for health, and research perspectives for innovative functional foods. Crit. Rev. Food Sci. Nutr. 54, 1584–1598 (2014).
Tomas-Barberan, F. A., Garcia-Villalba, R., Gonzalez-Sarrias, A., Selma, M. V. & Espin, J. C. Ellagic acid metabolism by human gut microbiota: consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status. J. Agr. Food Chem. 62, 6535–6538 (2014).
Garcia-Villalba, R., Beltran, D., Espin, J. C., Selma, M. V. & Tomas-Barberan, F. A. Time course production of urolithins from ellagic acid by human gut microbiota. J. Agr. Food Chem. 61, 8797–8806 (2013).
Selma, M. V., Beltran, D., Garcia-Villalba, R., Espin, J. C. & Tomas-Barberan, F. A. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. 5, 1779–1784 (2014).
Adlercreutz, H. Lignans and human health. Crit. Rev. Clin. Lab Sci. 44, 483–525 (2007).
Lampe, J. W. Is equol the key to the efficacy of soy foods? Am. J. Clin. Nutr. 89, 1664S–1667S (2009).
Patisaul, H. B. & Jefferson, W. The pros and cons of phytoestrogens. Front. Neuroendocrinol. 31, 400–419 (2010).
Setchell, K. D. & Clerici, C. Equol: history, chemistry, and formation. J. Nutr. 140, 1355S–1362S (2010).
Wu, A. H., Yu, M. C., Tseng, C. C. & Pike, M. C. Epidemiology of soy exposures and breast cancer risk. Br. J. Cancer 98, 9–14 (2008).
Setchell, K. D., Brown, N. M. & Lydeking-Olsen, E. The clinical importance of the metabolite equol-a clue to the effectiveness of soy and its isoflavones. J. Nutr. 132, 3577–3584 (2002).
Duncan, A. M., Merz-Demlow, B. E., Xu, X., Phipps, W. R. & Kurzer, M. S. Premenopausal equol excretors show plasma hormone profiles associated with lowered risk of breast cancer. Cancer Epidemiol. Biomarkers Prev. 9, 581–586 (2000).
Virk-Baker, M. K., Barnes, S., Krontiras, H. & Nagy, T. R. S-(−)equol producing status not associated with breast cancer risk among low isoflavone-consuming US postmenopausal women undergoing a physician-recommended breast biopsy. Nutr. Res. 34, 116–125 (2014).
Mazur, W. & Adlercreutz, H. Natural and anthropogenic environmental oestrogens: the scientific basis for risk assessment. Pure Appl. Chem. 70, 1759–1776 (1998).
Penalvo, J. L., Haajanen, K. M., Botting, N. & Adlercreutz, H. Quantification of lignans in food using isotope dilution gas chromatography/mass spectrometry. J. Agr. Food Chem. 53, 9342–9347 (2005).
Clavel, T., Borrmann, D., Braune, A., Dore, J. & Blaut, M. Occurrence and activity of human intestinal bacteria involved in the conversion of dietary lignans. Anaerobe 12, 140–147 (2006).
Mabrok, H. B. et al. Lignan transformation by gut bacteria lowers tumor burden in a gnotobiotic rat model of breast cancer. Carcinogenesis 33, 203–208 (2012). This study determines that the microbial production of bioactive metabolites from dietary sources mediates anticancer effects for the host.
Kaderlik, K. R. et al. Glucuronidation of N-hydroxy heterocyclic amines by human and rat liver microsomes. Carcinogenesis 15, 1695–1701 (1994).
Hirayama, K. et al. Effects of human intestinal flora on mutagenicity of and DNA adduct formation from food and environmental mutagens. Carcinogenesis 21, 2105–2111 (2000).
Kassie, F. et al. Intestinal microflora plays a crucial role in the genotoxicity of the cooked food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline. Carcinogenesis 22, 1721–1725 (2001).
Humblot, C. et al. β-glucuronidase in human intestinal microbiota is necessary for the colonic genotoxicity of the food-borne carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline in rats. Carcinogenesis 28, 2419–2425 (2007). This study shows that microbial glucuronidation activity in the gut contributes to the carcinogenic effect of heterocyclic amines from charred meat.
Craciun, S. & Balskus, E. P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl Acad. Sci. USA 109, 21307–21312 (2012).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011). This study demonstrates that the metabolism of dietary lipids by gut bacteria contributes to heart disease.
Tang, W. H. et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013).
Koeth, R. A. et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Drasar, B. S., Renwick, A. G. & Williams, R. T. The role of the gut flora in the metabolism of cyclamate. Biochem. J. 129, 881–890 (1972).
Legator, M. S., Palmer, K. A., Green, S. & Petersen, K. W. Cytogenetic studies in rats of cyclohexylamine, a metabolite of cyclamate. Science 165, 1139–1140 (1969).
Tamura, M., Hoshi, C. & Hori, S. Xylitol affects the intestinal microbiota and metabolism of daidzein in adult male mice. Int. J. Mol. Sci. 14, 23993–24007 (2013).
Suez, J. et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 514, 181–186 (2014).
Brown, R. J., de Banate, M. A. & Rother, K. I. Artificial sweeteners: a systematic review of metabolic effects in youth. Int. J. Pediatr. Obes. 5, 305–312 (2010).
Chassaing, B. et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015). This research shows that dietary supplements can disrupt mucus–bacterial interactions, promoting gut inflammation.
Hau, A. K., Kwan, T. H. & Li, P. K. Melamine toxicity and the kidney. J. Am. Soc. Nephrol. 20, 245–250 (2009).
Ingelfinger, J. R. Melamine and the global implications of food contamination. N. Engl. J. Med. 359, 2745–2748 (2008).
Zheng, X. et al. Melamine-induced renal toxicity is mediated by the gut microbiota. Sci. Transl. Med. 5, 172ra122 (2013). This study attributes the toxicity of dietary contaminants to gut microbial metabolism.
Shelton, D. R., Karns, J. S., McCarty, G. W. & Durham, D. R. Metabolism of melamine by Klebsiella terragena. Appl. Environ. Microbiol. 63, 2832–2835 (1997).
Jutzi, K., Cook, A. M. & Hutter, R. The degradative pathway of the s-triazine melamine. The steps to ring cleavage. Biochem. J. 208, 679–684 (1982).
Podschun, R. Isolation of Klebsiella terrigena from human feces: biochemical reactions, capsule types, and antibiotic sensitivity. Zentralbl. Bakteriol. 275, 73–78 (1991).
Sonnenburg, J. L. & Fischbach, M. A. Community health care: therapeutic opportunities in the human microbiome. Sci. Transl. Med. 3, 78ps12 (2011).
Ahmad, S. et al. A high throughput assay for discovery of bacterial β-glucuronidase inhibitors. Curr. Chem. Genom. 5, 13–20 (2011).
Ahmad, S., Hughes, M. A., Yeh, L. A. & Scott, J. E. Potential repurposing of known drugs as potent bacterial β-glucuronidase inhibitors. J. Biomol. Screen 17, 957–965 (2012).
Roberts, A. B., Wallace, B. D., Venkatesh, M. K., Mani, S. & Redinbo, M. R. Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol. Pharmacol. 84, 208–217 (2013).
Wallace, B. D. et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010). This is the first study to show that the toxicity associated with the microbial metabolism of cancer drugs in the gut can be mitigated using bacterial enzyme-specific small-molecule inhibitors.
LoGuidice, A., Wallace, B. D., Bendel, L., Redinbo, M. R. & Boelsterli, U. A. Pharmacologic targeting of bacterial β-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J. Pharmacol. Exp. Ther. 341, 447–454 (2012).
Carmody, R. N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015).
Lee, J. R. et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS ONE 10, e0122399 (2015).
Hooper, L. V., Littman, D. R. & Macpherson, A. J. Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012).
Viaud, S. et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013).
Viaud, S. et al. Cyclophosphamide induces differentiation of TH17 cells in cancer patients. Cancer Res. 71, 661–665 (2011).
Iida, N. et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970 (2013).
Ozben, T. Oxidative stress and apoptosis: impact on cancer therapy. J. Pharm. Sci. 96, 2181–2196 (2007).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Vetizou, M. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015).
Garrett, W. S. et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131, 33–45 (2007).
Neurath, M. F. New targets for mucosal healing and therapy in inflammatory bowel diseases. Mucosal Immunol. 7, 6–19 (2014).
Rooks, M. G. et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 8, 1403–1417 (2014).
Donia, M. S. et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414 (2014). This study reveals the potential of the human microbiome as a rich source of bioactive natural products, including antibiotics.
Rupnik, M., Wilcox, M. H. & Gerding, D. N. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7, 526–536 (2009).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Sorg, J. A. & Sonenshein, A. L. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 192, 4983–4990 (2010).
Wilson, K. H. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 18, 1017–1019 (1983).
Acknowledgements
The authors apologize to all of those colleagues whose work could not be included in this Review owing to space constraints. The authors also thank the reviewers for their comments and suggestions. This work was supported by the US National Institutes of Health (R01AT008618, R01HL122593 and F32DK101154), the Young Investigator Grant for Probiotics Research, the George Williams Hooper Research Foundation and the University of California San Francisco (UCSF) Department of Microbiology & Immunology. P.J.T. is a Nadia's Gift Foundation Innovator supported, in part, by the Damon Runyon Cancer Research Foundation (DRR-42-16).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
P.J.T. is on the Scientific Advisory Board for Seres Therapeutics and Whole Biome, has consulted for Pfizer in the past year and has current research support from MedImmune.
Supplementary information
Supplementary information
Comprehensive list of pharmaceuticals and dietary compounds subject to gut microbial metabolism (PDF 591 kb)
Glossary
- Microbiome
-
The combined genetic material and metabolic activities of the microbiota.
- Microbiota
-
The collection of all microorganisms (archaea, bacteria, microscopic fungi, parasites and viruses) found in a given body habitat.
- Xenobiotics
-
Compounds that are foreign to a biological system. For humans, these include drugs, dietary bioactive compounds, food additives and environmental toxins.
- Azo bond
-
A chemical bond composed of N=N.
- Pharmacogenetics
-
The study of how genetic factors influence therapeutic outcomes.
- Pharmacogenomics
-
The use of sequencing-based genomic methods to analyse the links between genetics and therapeutic outcomes.
- Bioavailability
-
The proportion of an administered compound that reaches systemic circulation and thus has the potential to influence the intended target.
- First-pass metabolism
-
The metabolism of orally ingested compounds before reaching general circulation.
- Biliary excretion
-
The transfer of xenobiotics and other compounds from the plasma to bile through hepatocytes, which is followed by the release of the compounds into the gut lumen.
- Enterohepatic circulation
-
The circulation of xenobiotics and endogenous compounds that are absorbed from the intestines, transported to the liver, and then re-enter the intestine through the bile ducts, where they may be reabsorbed or metabolized by the gut microbiota.
- Reduction
-
A chemical reaction in which the oxidation state of a chemical bond is reduced. For example, a carbon–carbon bond modified to a carbon–hydrogen bond is a reductive transformation.
- Hydrolysis
-
A chemical reaction in which a chemical bond is cleaved using a water molecule, which acts as the nucleophile.
- Cytochrome P450 enzymes
-
(CYPs) A family of enzymes that is responsible for the oxidative biotransformation of xenobiotics and other compounds.
- Prodrugs
-
Drugs that are administered in an inactive form and become active when metabolized.
- Folate
-
A B vitamin that is essential for DNA synthesis, DNA repair and other biological reactions.
- Germ-free
-
Animals devoid of microorganisms.
- Gnotobiotic
-
The colonization of germ-free animals with individual microorganisms or defined microbial communities.
- Glucuronidation
-
The addition of glucuronic acid to a substrate. Glucuronidation is used as a mechanism of xenobiotic metabolism by the host.
- Bile acids
-
Steroid acids produced by the liver that emulsify fats during digestion.
- Aglycone
-
The remaining compound after the removal of a glycosyl moiety.
- Serum metabolome
-
The collection of all metabolites found in serum.
- Conjugation
-
The addition of a chemical unit (for example, glucuronic acid or glutathione) to xenobiotics, increasing the solubility and molecular weight of the parent compound and facilitating elimination from the body.
- Metabolic syndrome
-
A collection of physiological and biochemical conditions, defined as a combination of high blood pressure, increased blood sugar levels, excess fat and abnormal cholesterol levels. This syndrome increases the risk of heart disease, stroke and diabetes.
- Metformin
-
An oral medication used to treat type 2 diabetes.
- Pharmacopoeia
-
A manual for the preparation and use of medicinal drugs. The name is derived from the Greek words pharmakon (drug) and -poios (making).
Rights and permissions
About this article
Cite this article
Spanogiannopoulos, P., Bess, E., Carmody, R. et al. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 14, 273–287 (2016). https://doi.org/10.1038/nrmicro.2016.17
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2016.17
This article is cited by
-
Gut microbiota, circulating cytokines and dementia: a Mendelian randomization study
Journal of Neuroinflammation (2024)
-
Gut microbiome modulates tacrolimus pharmacokinetics through the transcriptional regulation of ABCB1
Microbiome (2023)
-
Parkinson’s disease and gut microbiota: from clinical to mechanistic and therapeutic studies
Translational Neurodegeneration (2023)
-
Genome-scale metabolic reconstruction of 7,302 human microorganisms for personalized medicine
Nature Biotechnology (2023)
-
Sexual Dimorphic Interplays Between Gut Microbiota and Antihypertensive Drugs
Current Hypertension Reports (2023)