Key Points
-
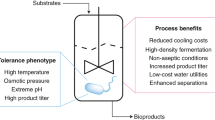
One of the pillars of contemporary synthetic biology is the use of reliable biological chassis into which users can plug-in and plug-out genetic circuits and new-to-nature properties at will.
-
The microorganisms that are easiest to genetically manipulate in the laboratory are frequently suboptimal for industrial applications owing to physicochemical stress and harsh operation conditions.
-
Hallmark features of pseudomonads as synthetic biology platforms include their pre-endowed metabolic, physiological and stress-endurance traits, which make them adequate for real-life biotechnological needs.
-
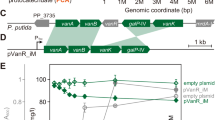
The range of molecular tools that are available for the rational manipulation of pseudomonads is continuously increasing in variety and scope. These assets include streamlined plasmid vectors for gene expression and genome editing, as well as a number of in silico tools for the modelling and prediction of metabolic and physiological behaviour of these bacteria.
-
String–weight biological engineering enables the assembly of complex biosystems by combining the physical connectivity of available parts and modules with evolutionary gravitation of input–output transfer functions towards functional optimality.
Abstract
Much of contemporary synthetic biology research relies on the use of bacterial chassis for plugging-in and plugging-out genetic circuits and new-to-nature functionalities. However, the microorganisms that are the easiest to manipulate in the laboratory are often suboptimal for downstream industrial applications, which can involve physicochemical stress and harsh operating conditions. In this Review, we advocate the use of environmental Pseudomonas strains as model organisms that are pre-endowed with the metabolic, physiological and stress-endurance traits that are demanded by current and future synthetic biology and biotechnological needs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
McPherson, M. J. & Parish, J. H. Applications of recombinant DNA in biotechnology. Nat. Prod. Rep. 4, 205–224 (1987).
Malik, V. S. Biotechnology — the golden age. Adv. Appl. Microbiol. 34, 263–306 (1989).
Ramos, J. L. et al. The behavior of bacteria designed for biodegradation. Biotechnology 12, 1349–1356 (1994).
Pieper, D. H. & Reineke, W. Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 11, 262–270 (2000).
Janssen, D. B., Dinkla, I. J., Poelarends, G. J. & Terpstra, P. Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities. Environ. Microbiol. 7, 1868–1882 (2005).
Ferrer, M., Beloqui, A., Timmis, K. N. & Golyshin, P. N. Metagenomics for mining new genetic resources of microbial communities. J. Mol. Microbiol. Biotechnol. 16, 109–123 (2009).
Dietrich, J. A., McKee, A. E. & Keasling, J. D. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu. Rev. Biochem. 79, 563–590 (2010).
Galvão, T. C., Mohn, W. W. & de Lorenzo, V. Exploring the microbial biodegradation and biotransformation gene pool. Trends Biotechnol. 23, 497–506 (2005).
Tuffin, M., Anderson, D., Heath, C. & Cowan, D. A. Metagenomic gene discovery: how far have we moved into novel sequence space? Biotechnol. J. 4, 1671–1683 (2009).
Purnick, P. E. & Weiss, R. The second wave of synthetic biology: from modules to systems. Nature Rev. Mol. Cell. Biol. 10, 410–422 (2009).
Keasling, J. D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 3, 64–76 (2008).
Endy, D. Foundations for engineering biology. Nature 438, 449–453 (2005). This paper makes a seminal contribution to the synthetic biology field and sets the basis for many of the current tenets within the discipline.
Danchin, A. Scaling up synthetic biology: do not forget the chassis. FEBS Lett. 586, 2129–2137 (2012). This paper provides an enlightening discussion on the neglected issue of choosing an appropriate chassis for synthetic biology applications.
de Lorenzo, V. Beware of metaphors: chasses and orthogonality in synthetic biology. Bioeng. Bugs 2, 3–7 (2011).
Wheeler, M. B. & Campion, D. R. Animal production — a longstanding biotechnological success. Am. J. Clin. Nutr. 58, (Suppl. 2) 276S–281S (1993).
Sauer, M. & Mattanovich, D. Construction of microbial cell factories for industrial bioprocesses. J. Chem. Technol. Biotechnol. 87, 445–450 (2012).
Schmid, A. et al. Industrial biocatalysis today and tomorrow. Nature 409, 258–268 (2001).
Foley, P. L. & Shuler, M. L. Considerations for the design and construction of a synthetic platform cell for biotechnological applications. Biotechnol. Bioeng. 105, 26–36 (2010).
Dürre, P. New insights and novel developments in clostridial acetone/butanol/isopropanol fermentation. Appl. Microbiol. Biotechnol. 49, 639–648 (1998).
Karlsson, E. N., Johansson, L., Holst, O. & Lidén, G. in The Metabolic Pathway Engineering Handbook: Fundamentals (ed. Smolke, C.D) 21.21–21.23 (CRC Press, 2010).
Copley, S. D. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nature Chem. Biol. 5, 559–566 (2009).
de Lorenzo, V. Systems biology approaches to bioremediation. Curr. Opin. Biotechnol. 19, 579–589 (2008).
Kivisaar, M. Mechanisms of stationary-phase mutagenesis in bacteria: mutational processes in pseudomonads. FEMS Microbiol. Lett. 312, 1–14 (2010).
Pérez-Pantoja, D., Nikel, P. I., Chavarría, M. & de Lorenzo, V. Endogenous stress caused by faulty oxidation reactions fosters evolution of 2,4-dinitrotoluene-degrading bacteria. PLoS Genet. 9, e1003764 (2013). This article reveals the role of oxidative stress caused by the biological oxidation of aromatic compounds as the driving force for the evolution of biodegradation pathways.
Nikel, P. I., Pérez-Pantoja, D. & de Lorenzo, V. Why are chlorinated pollutants so difficult to degrade aerobically? Redox stress limits 1,3-dichloroprop-1-ene metabolism by Pseudomonas pavonaceae. Phil. Trans. R. Soc. B 368, 20120377 (2013).
Imlay, J. A. in EcoSal — Escherichia coli and Salmonella: Cellular and Molecular Biology (ed. Böck, A. et al.) (ASM Press, 2009).
Euzéby, J. P. List of bacterial names with standing in nomenclature: a folder available on the internet. Internatl J. Syst. Evol. Microbiol. 47, 590–592 (1997).
Palleroni, N. J. in Bergey's Manual of Systematic Bacteriology (ed. Kreig, N. R.) 141–199 (Williams & Wilkins, 1984).
Palleroni, N. J. The Pseudomonas story. Environ. Microbiol. 12, 1377–1383 (2010).
Peix, A., Ramírez-Bahena, M. H. & Velázquez, E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 9, 1132–1147 (2009).
Gurney, R. & Thomas, C. M. Mupirocin: biosynthesis, special features and applications of an antibiotic from a Gram-negative bacterium. Appl. Microbiol. Biotechnol. 90, 11–21 (2011).
Loper, J. E. et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8, e1002784 (2012).
Balasubramanian, D., Schneper, L., Kumari, H. & Mathee, K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 41, 1–20 (2013).
Rodríguez-Rojas, A., Oliver, A. & Blázquez, J. Intrinsic and environmental mutagenesis drive diversification and persistence of Pseudomonas aeruginosa in chronic lung infections. J. Infect. Dis. 205, 121–127 (2012).
Fuhrer, T., Fischer, E. & Sauer, U. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187, 1581–1590 (2005).
May, T. B. & Chakrabarty, A. M. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 2, 151–157 (1994).
de Lorenzo, V. From the selfish gene to selfish metabolism: revisiting the central dogma. BioEssays. 36, 226–235 (2014).
Oberhardt, M. A., Puchałka, J., Fryer, K. E., Martins dos Santos, V. A. P. & Papin, J. A. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J. Bacteriol. 190, 2790–2803 (2008).
Blank, L. M., Ionidis, G., Ebert, B. E., Bühler, B. & Schmid, A. Metabolic response of Pseudomonas putida during redox biocatalysis in the presence of a second octanol phase. FEBS J. 275, 5173–5190 (2008).
Aires, J. R., Köhler, T., Nikaido, H. & Plésiat, P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43, 2624–2628 (1999).
Vettoretti, L. et al. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 53, 1987–1997 (2009).
Udaondo, Z. et al. Analysis of solvent tolerance in Pseudomonas putida DOT-T1E based on its genome sequence and a collection of mutants. FEBS Lett. 586, 2932–2938 (2012).
Timmis, K. N. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ. Microbiol. 4, 779–781 (2002).
Poblete-Castro, I., Becker, J., Dohnt, K., Martins dos Santos, V. A. P. & Wittmann, C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 93, 2279–2290 (2012).
Kevles, D. J. Ananda Chakrabarty wins a patent: biotechnology, law, and society. Hist. Stud. Phys. Biol. Sci. 25, 111–135 (1994).
Wu, X. et al. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 35, 299–323 (2011).
Matilla, M. A. et al. Complete genome of the plant growth-promoting rhizobacterium Pseudomonas putida BIRD-1. J. Bacteriol. 193, 1290 (2011).
Roca, A. et al. Analysis of the plant growth-promoting properties encoded by the genome of the rhizobacterium Pseudomonas putida BIRD-1. Environ. Microbiol. 15, 780–794 (2013).
Silby, M. W., Winstanley, C., Godfrey, S. A., Levy, S. B. & Jackson, R. W. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 35, 652–680 (2011).
Jiménez, J. I., Miñambres, B., García, J. L. & Díaz, E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4, 824–841 (2002). A comprehensive survey of the many biochemical mechanisms that enable P. putida to grow on diverse carbon substrates.
Mohn, W. W., Garmendia, J., Galvão, T. C. & de Lorenzo, V. Surveying biotransformations with à la carte genetic traps: translating dehydrochlorination of lindane (γ-hexachlorocyclohexane) into lacZ-based phenotypes. Environ. Microbiol. 8, 546–555 (2006).
Orphan, V. J. Methods for unveiling cryptic microbial partnerships in nature. Curr. Opin. Microbiol. 12, 231–237 (2009).
Watrous, J. et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl Acad. Sci. USA 109, E1743–E1752 (2012).
Bacchus, W. & Fussenegger, M. Engineering of synthetic intercellular communication systems. Metab. Eng. 16, 33–41 (2013).
Ishige, T., Honda, K. & Shimizu, S. Whole organism biocatalysis. Curr. Opin. Chem. Biol. 9, 174–180 (2005).
Inoue, A. & Horikoshi, K. A. Pseudomonas thrives in high concentrations of toluene. Nature 227, 264–265 (1989). This is the very first report of the isolation of a Pseudomonas species that is highly tolerant to solvents.
Ramos, J. L., Duque, E., Huertas, M. J. & Haidour, A. Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177, 3911–3916 (1995).
Weber, F. J., Ooijkaas, L. P., Schemen, R. M., Hartmans, S. & de Bont, J. A. Adaptation of Pseudomonas putida S12 to high concentrations of styrene and other organic solvents. Appl. Environ. Microbiol. 59, 3502–3504 (1993).
Ramos, J. L. et al. Mechanisms of solvent tolerance in Gram-negative bacteria. Annu. Rev. Microbiol. 56, 743–768 (2002).
Domínguez-Cuevas, P., González-Pastor, J. E., Marqués, S., Ramos, J. L. & de Lorenzo, V. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J. Biol. Chem. 281, 11981–11991 (2006).
Rühl, J., Schmid, A. & Blank, L. M. Selected Pseudomonas putida strains able to grow in the presence of high butanol concentrations. Appl. Environ. Microbiol. 75, 4653–4656 (2009).
Nielsen, D. R. et al. Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 11, 262–273 (2009).
Luengo, J. M., García, B., Sandoval, A., Naharro, G. & Olivera, E. R. Bioplastics from microorganisms. Curr. Opin. Microbiol. 6, 251–260 (2003).
Anderson, A. J. & Dawes, E. A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54, 450–472 (1990).
Keshavarz, T. & Roy, I. Polyhydroxyalkanoates: bioplastics with a green agenda. Curr. Opin. Microbiol. 13, 321–326 (2010).
Nikel, P. I., de Almeida, A., Melillo, E. C., Galvagno, M. A. & Pettinari, M. J. New recombinant Escherichia coli strain tailored for the production of poly(3-hydroxybutyrate) from agroindustrial by-products. Appl. Environ. Microbiol. 72, 3949–3954 (2006).
Gao, X., Chen, J. C., Wu, Q. & Chen, G. Q. Polyhydroxyalkanoates as a source of chemicals, polymers, and biofuels. Curr. Opin. Biotechnol. 22, 768–774 (2011).
Kim, Y., Kim, H. W., Chung, M. G. & Rhee, Y. H. Biosynthesis, modification, and biodegradation of bacterial medium-chain-length polyhydroxyalkanoates. J. Microbiol. 45, 87–97 (2007).
Arias, S., Bassas-Galia, M., Molinari, G. & Timmis, K. N. Tight coupling of polymerization and depolymerization of polyhydroxyalkanoates ensures efficient management of carbon resources in Pseudomonas putida. Microb. Biotechnol. 6, 551–563 (2013).
Tripathi, L. et al. Pseudomonas putida KT2442 as a platform for the biosynthesis of polyhydroxyalkanoates with adjustable monomer contents and compositions. Bioresour. Technol. 142, 225–231 (2013).
Escapa, I. F. et al. Disruption of β-oxidation pathway in Pseudomonas putida KT2442 to produce new functionalized PHAs with thioester groups. Appl. Microbiol. Biotechnol. 89, 1583–1598 (2011).
Regenhardt, D. et al. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 4, 912–915 (2002).
Nakazawa, T. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4, 782–786 (2002).
Feist, C. F. & Hegeman, G. D. Regulation of the meta cleavage pathway for benzoate oxidation by Pseudomonas putida. J. Bacteriol. 100, 1121–1123 (1969).
Williams, P. A. & Murray, K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J. Bacteriol. 120, 416–423 (1974).
Federal Register. Appendix E, Certified host-vector systems. 47, 17197 (1982).
Nelson, K. E. et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4, 799–808 (2002). This paper reports the completion of the genome sequencing that enabled the understanding and rational exploration of the metabolic diversity and physiological properties of strain KT2440.
Martins dos Santos, V. A. P., Heim, S., Moore, E. R., Strätz, M. & Timmis, K. N. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 6, 1264–1286 (2004).
Chavarría, M., Nikel, P. I., Pérez-Pantoja, D. & de Lorenzo, V. The Entner–Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ. Microbiol. 15, 1772–1785 (2013).
Ebert, B. E., Kurth, F., Grund, M., Blank, L. M. & Schmid, A. Response of Pseudomonas putida KT2440 to increased NADH and ATP demand. Appl. Environ. Microbiol. 77, 6597–6605 (2011).
Nogales, J., Palsson, B. Ø. & Thiele, I. A genome-scale metabolic reconstruction of Pseudomonas putida KT2440: iJN746 as a cell factory. BMC Syst. Biol. 2, 79 (2008). This paper reports on the development of the first genome-wide metabolic model of strain KT2440, which has enabled the identification of this bacterium as a useful microbial cell factory.
Puchałka, J. et al. Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput. Biol. 4, e1000210 (2008).
Sohn, S. B., Kim, T. Y., Park, J. M. & Lee, S. Y. In silico genome-scale metabolic analysis of Pseudomonas putida KT2440 for polyhydroxyalkanoate synthesis, degradation of aromatics and anaerobic survival. Biotechnol. J. 5, 739–750 (2010).
van Duuren, J. B. et al. Reconciling in vivo and in silico key biological parameters of Pseudomonas putida KT2440 during growth on glucose under carbon-limited condition. BMC Biotechnol. 13, 93 (2013).
Nikel, P. I., Kim, J. & de Lorenzo, V. Metabolic and regulatory rearrangements underlying glycerol metabolism in Pseudomonas putida KT2440. Environ. Microbiol. 16, 239–254 (2014).
Kim, J., Oliveros, J. C., Nikel, P. I., de Lorenzo, V. & Silva-Rocha, R. Transcriptomic fingerprinting of Pseudomonas putida under alternative physiological regimes. Environ. Microbiol. Rep. 5, 883–891 (2013).
Park, S. J. et al. PutidaNET: interactome database service and network analysis of Pseudomonas putida KT2440. BMC Genomics 10, S18 (2009).
Winsor, G. L. et al. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600 (2011).
Schweizer, H. P. Vectors to express foreign genes and techniques to monitor gene expression in Pseudomonads. Curr. Opin. Biotechnol. 12, 439–445 (2001).
Tavita, K. et al. Homologous recombination is facilitated in starving populations of Pseudomonas putida by phenol stress and affected by chromosomal location of the recombination target. Mutat. Res. 737, 12–24 (2012).
Kolisnychenko, V. et al. Engineering a reduced Escherichia coli genome. Genome Res. 12, 640–647 (2002).
Martínez-García, E. & de Lorenzo, V. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ. Microbiol. 13, 2702–2716 (2011). This paper presents a method for the seamless deletion of genetic elements in Pseudomonas spp. and other Gram-negative bacteria.
Martínez-García, E. & de Lorenzo, V. Transposon-based and plasmid-based genetic tools for editing genomes of Gram-negative bacteria. Methods Mol. Biol. 813, 267–283 (2012).
Martínez-García, E., Nikel, P. I., Chavarría, M. & de Lorenzo, V. The metabolic cost of flagellar motion in Pseudomonas putida KT2440. Environ. Microbiol. 16, 291–303 (2014).
LePrince, A., de Lorenzo, V., Voller, P., van Passel, M. W. & Martins dos Santos, V. A. P. Random and cyclical deletion of large DNA segments in the genome of Pseudomonas putida. Environ. Microbiol. 14, 1444–1453 (2012).
Bagdasarian, M. et al. Specific purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16, 237–247 (1981).
Nikel, P. I. & de Lorenzo, V. Engineering an anaerobic metabolic regime in Pseudomonas putida KT2440 for the anoxic biodegradation of 1,3-dichloroprop-1-ene. Metab. Eng. 15, 98–112 (2013). This paper is a practical example of how synthetic biology tools can be used to change the entire lifestyle of a bacterium, in this case, moving the metabolism of P. putida towards anaerobiosis.
de Lorenzo, V. & Timmis, K. N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235, 386–405 (1994).
Martínez-García, E., Calles, B., Arévalo-Rodríguez, M. & de Lorenzo, V. pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol. 11, 38 (2011).
Nikel, P. I. & de Lorenzo, V. Implantation of unmarked regulatory and metabolic modules in Gram-negative bacteria with specialised mini-transposon delivery vectors. J. Biotechnol. 163, 143–154 (2013).
Vora, T., Hottes, A. K. & Tavazoie, S. Protein occupancy landscape of a bacterial genome. Mol. Cell 35, 247–253 (2009).
Sousa, C., de Lorenzo, V. & Cebolla, A. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology 143, 2071–2078 (1997).
Choi, K. H. & Schweizer, H. P. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nature Protocols 1, 153–161 (2006).
Lambertsen, L., Sternberg, C. & Molin, S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ. Microbiol. 6, 726–732 (2004).
de las Heras, A., Carreño, C. A. & de Lorenzo, V. Stable implantation of orthogonal sensor circuits in Gram-negative bacteria for environmental release. Environ. Microbiol. 10, 3305–3316 (2008).
Miyazaki, R. & van der Meer, J. R. A new large-DNA-fragment delivery system based on integrase activity from an integrative and conjugative element. Appl. Environ. Microbiol. 79, 4440–4447 (2013).
Cases, I. & de Lorenzo, V. Promoters in the environment: transcriptional regulation in its natural context. Nature Rev. Microbiol. 3, 105–118 (2005).
Blatny, J. M., Brautaset, T., Winther-Larsen, H. C., Haugan, K. & Valla, S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63, 370–379 (1997).
de Lorenzo, V., Fernández, S., Herrero, M., Jakubzik, U. & Timmis, K. N. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene 130, 41–46 (1993).
Panke, S., de Lorenzo, V., Kaiser, A., Witholt, B. & Wubbolts, M. G. Engineering of a stable whole-cell biocatalyst capable of (S)-styrene oxide formation for continuous two-liquid-phase applications. Appl. Environ. Microbiol. 65, 5619–5623 (1999).
Silva-Rocha, R., de Jong, H., Tamames, J. & de Lorenzo, V. The logic layout of the TOL network of Pseudomonas putida pWW0 plasmid stems from a metabolic amplifier motif (MAM) that optimizes biodegradation of m-xylene. BMC Syst. Biol. 5, 191 (2011).
Billerbeck, S., Calles, B., Müller, C. L., de Lorenzo, V. & Panke, S. Towards functional orthogonalization of protein complexes: individualization of GroEL monomers leads to distinct quasi-homogeneous single rings. ChemBiochem 14, 2310–2321 (2013).
Calles, B. & de Lorenzo, V. Expanding the Boolean logic of the prokaryotic transcription factor XylR by functionalization of permissive sites with a protease-target sequence. ACS Synth. Biol. 2, 594–603 (2013).
Kochanowski, K. et al. Functioning of a metabolic flux sensor in Escherichia coli. Proc. Natl Acad. Sci. USA 110, 1130–1135 (2013).
Garmendia, J., de las Heras, A., Galvão, T. C. & de Lorenzo, V. Tracing explosives in soil with transcriptional regulators of Pseudomonas putida evolved for responding to nitrotoluenes. Microb. Biotechnol. 1, 236–246 (2008).
Zhang, C., Wei, Z. H. & Ye, B. C. Quantitative monitoring of 2-oxoglutarate in Escherichia coli cells by a fluorescence resonance energy transfer-based biosensor. Appl. Microbiol. Biotechnol. 97, 8307–8316 (2013).
de las Heras, A., Carreño, C. A., Martínez-García, E. & de Lorenzo, V. Engineering input/output nodes in prokaryotic regulatory circuits. FEMS Microbiol. Rev. 34, 842–865 (2010).
Basu, S., Gerchman, Y., Collins, C. H., Arnold, F. H. & Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005).
Silva-Rocha, R. & de Lorenzo, V. Engineering multicellular logic in bacteria with metabolic wires. ACS Synth. Biol. http://dx.doi.org/10.1021/sb400064y (2014).
Gross, R., Buehler, K. & Schmid, A. Engineered catalytic biofilms for continuous large scale production of n-octanol and (S)-styrene oxide. Biotechnol. Bioeng. 110, 424–436 (2013).
Pósfai, G. et al. Emergent properties of reduced-genome Escherichia coli. Science 312, 1044–1046 (2006).
Ma, S., Tang, N. & Tian, J. DNA synthesis, assembly and applications in synthetic biology. Curr. Opin. Chem. Biol. 16, 260–267 (2012).
Huerta, S. Structural design in the work of Gaudí. Archit. Sci. Rev. 49, 324–339 (2006).
Ben Yehezkel, T., Biezuner, T., Linshiz, G., Mazor, Y. & Shapiro, E. Programmable in vivo selection of arbitrary DNA sequences. PLoS ONE 7, e47795 (2012).
Oliver, A., Cantón, R., Campo, P., Baquero, F. & Blázquez, J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288, 1251–1254 (2000).
van Ham, R. C. et al. Reductive genome evolution in Buchnera aphidicola. Proc. Natl Acad. Sci. USA 100, 581–586 (2003).
Chou, H. H. & Keasling, J. D. Programming adaptive control to evolve increased metabolite production. Nature Comm. 4, 2595 (2013).
Shetty, R. P., Endy, D. & Knight, T. F. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2, 5 (2008).
Anderson, J. C. et al. BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 4, 1 (2010).
Silva-Rocha, R. et al. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675 (2013). Building on the standardization premise of synthetic biology, the authors of this paper propose the adoption of a universal framework for plasmid vectors.
Pfleger, B. F., Pitera, D. J., Smolke, C. D. & Keasling, J. D. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nature Biotech. 24, 1027–1032 (2006).
Santos, C. N. S. & Stephanopoulos, G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr. Opin. Chem. Biol. 12, 168–176 (2008).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009).
Agapakis, C. M. & Silver, P. A. Synthetic biology: exploring and exploiting genetic modularity through the design of novel biological networks. Mol. BioSyst. 5, 704–713 (2009).
Jayanthi, S., Nilgiriwala, K. S. & Del Vecchio, D. Retroactivity controls the temporal dynamics of gene transcription. ACS Synth. Biol. 2, 431–441 (2013).
Chavarría, M., Kleijn, R. J., Sauer, U., Pflüger-Grau, K. & de Lorenzo, V. Regulatory tasks of the phosphoenolpyruvate-phosphotransferase system of Pseudomonas putida in central carbon metabolism. mBio 3, e00028-12 (2012).
Acknowledgements
This study was supported by the BIO and FEDER CONSOLIDER-INGENIO Program of the Spanish Ministry of Science and Innovation, the ST-FLOW, ARYSIS and EVOPROG Contracts of the EU, the ERANET-IB Program, and the PROMT Project of the CAM. PIN is a researcher from the Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina) and holds a Marie Curie Actions Programme grant obtained from the EU (ALLEGRO, UE-FP7-PEOPLE-2011-IIF-300508).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Forward-engineering
-
The rational design of complex objects with properties that can be quantitatively predicted from the attributes of the components.
- Biological chassis
-
An autonomous genetic and/or biochemical scaffold that functions as an dynamic platform for implanting forward-designed biological devices.
- Logic gates
-
Devices that execute binary, Boolean operations in which (typically) one or two inputs with values 1 or 0 are processed into one fixed, digital output (0 or 1 as well). The output of one gate can become the input of another gate, thereby enabling composability and scalability.
Rights and permissions
About this article
Cite this article
Nikel, P., Martínez-García, E. & de Lorenzo, V. Biotechnological domestication of pseudomonads using synthetic biology. Nat Rev Microbiol 12, 368–379 (2014). https://doi.org/10.1038/nrmicro3253
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro3253
This article is cited by
-
Genome-scale metabolic network model and phenome of solvent-tolerant Pseudomonas putida S12
BMC Genomics (2024)
-
Increasing cellular fitness and product yields in Pseudomonas putida through an engineered phosphoketolase shunt
Microbial Cell Factories (2023)
-
Insights into the susceptibility of Pseudomonas putida to industrially relevant aromatic hydrocarbons that it can synthesize from sugars
Microbial Cell Factories (2023)
-
Optimized enantioselective (S)-2-hydroxypropiophenone synthesis by free- and encapsulated-resting cells of Pseudomonas putida
Microbial Cell Factories (2023)
-
Mechanism of furfural toxicity and metabolic strategies to engineer tolerance in microbial strains
Microbial Cell Factories (2023)