Key Points
-
Many marine sponges (phylum Porifera), the most ancient of the metazoan animals, contain dense and diverse microbial communities.
-
Members of ∼30 bacterial phyla and several archaeal lineages have been reported to contribute to the enormous microbial diversity in marine sponges. The symbiotic microbial consortia are located extracellularly in the mesohyl matrix. Transmission to the next generation occurs vertically through the reproductive stages, but horizontal transmission might also be possible. The mechanisms by which microbial diversity in sponges is shaped and maintained throughout the sponge life cycle, as well as through evolutionary time, are the subject of much current debate.
-
The metabolic capabilities of sponge-associated microorganisms are becoming increasingly well understood, largely as a result of metagenomic, metaproteogenomic and single-cell genomics studies. Furthermore, several putative symbiosis factors, such as proteins that contain eukaryotic domains (that is, ankyrin repeats, tetratrico peptide repeats and leucine-rich repeats), have been identified.
-
The genome of the sponge Amphimedon queenslandica provides new insights into metazoan evolution and also adds a new angle to investigating the mechanisms of sponge–microorganism interactions. Of particular interest are the pattern recognition receptors of the innate immune system, which recognize microbial ligands.
-
Metagenomic and single-cell genomics approaches are promising for the field of marine drug development, as they provide biotechnological access to pharmacologically important host- and symbiont-derived compounds.
-
Sponges represent an important and tractable model system for the study of metazoan evolution, host–microorganism interactions and chemical diversity.
Abstract
Marine sponges (phylum Porifera) often contain dense and diverse microbial communities, which can constitute up to 35% of the sponge biomass. The genome of one sponge, Amphimedon queenslandica, was recently sequenced, and this has provided new insights into the origins of animal evolution. Complementary efforts to sequence the genomes of uncultivated sponge symbionts have yielded the first glimpse of how these intimate partnerships are formed. The remarkable microbial and chemical diversity of the sponge–microorganism association, coupled with its postulated antiquity, makes sponges important model systems for the study of metazoan host–microorganism interactions, and their evolution, as well as for enabling access to biotechnologically important symbiont-derived natural products. In this Review, we discuss our current understanding of the interactions between marine sponges and their microbial symbiotic consortia, and highlight recent insights into these relationships from genomic studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bell, J. J. The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 79, 341–353 (2008).
Dayton, P. K. Interdecadal variation in an antarctic sponge and its predators from oceanographic climate shifts. Science 245, 1484–1486 (1989).
Bell, J. J. & Smith, D. Ecology of sponge assemblages (Porifera) in the Wakatobi region, south-east Sulawesi, Indonesia: richness and abundance. J. Mar. Biol. Assoc. UK 84, 581–591 (2004).
Brusca, R. C. & Brusca, G. J. Invertebrates 2nd edn (Sinauer Associates, 2003).
Pawlik, J. R. The chemical ecology of sponges on Caribbean reefs: natural products shape natural systems. Bioscience 61, 888–898 (2011).
Li, C. W., Chen, J. Y. & Hua, T. E. Precambrian sponges with cellular structures. Science 279, 879–882 (1998).
Love, G. D. et al. Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 457, 718–721 (2009).
Taylor, M. W., Radax, R., Steger, D. & Wagner, M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347 (2007). An excellent, comprehensive review of sponge microbiology.
Vacelet, J. & Donadey, C. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30, 301–314 (1977).
Wilkinson, C. R., Garrone, R. & Vacelet, J. Marine sponges discriminate between food bacteria and bacterial symbionts: electron microscope radioautography and in situ evidence. Proc. R. Soc. Lond. B 220, 519–528 (1984). An early paper in sponge microbiology that is noteworthy for its thought-provoking hypotheses, which are stimulating contemporary discussions.
Wehrl, M., Steinert, M. & Hentschel, U. Bacterial uptake by the marine sponge Aplysina aerophoba. Microb. Ecol. 53, 355–365 (2007).
Müller, W. E. G. & Müller, I. M. Origin of the metazoan immune system: identification of the molecules and their functions in sponges. Integr. Comp. Biol. 43, 281–292 (2003).
Wiens, M. et al. Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol. Biol. Evol. 24, 792–804 (2007).
Srivastava, M. et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 (2010). The first, and still only, complete genome from a sponge reveals that most gene families which are characteristic of true animals, including humans, were already present in the last common ancestor of all animals.
Gauthier, E. A., Du Pasquier, L. & Degnan, B. M. The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of Toll-like and interleukin 1 receptor pathways. Evol. Dev. 12, 519–533 (2010).
Blunt, J. W., Copp, B. R., Munro, M. H. G., Northcote, P. T. & Prinsep, M. R. Marine natural products. Nat. Prod. Rep. 28, 196–268 (2011).
Piel, J. Bacterial symbionts: prospects for the sustainable production of invertebrate-derived pharmaceuticals. Curr. Med. Chem. 13, 39–50 (2006).
Sipkema, D. et al. Multiple approaches to enhance the cultivability of bacteria associated with the marine sponge Haliclona (gellius) sp. Appl. Environ. Microbiol. 77, 2130–2140 (2011).
Bright, M. & Bulgheresi, S. A complex journey: transmission of microbial symbionts. Nature Rev. Microbiol. 8, 218–230 (2010).
Wilson, A. C. et al. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 19, 249–258 (2010).
Dale, C. & Moran, N. A. Molecular interactions between bacterial symbionts and their hosts. Cell 126, 453–465 (2006).
Nyholm, S. V. & McFall-Ngai, M. J. The winnowing: establishing the squid–Vibrio symbiosis. Nature Rev. Microbiol. 2, 632–642 (2004). A brilliant example of the intricate interactions that can evolve between animal host and bacterial symbiont.
Schmitt, S., Angermeier, H., Schiller, R., Lindquist, N. & Hentschel, U. Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 74, 7694–7708 (2008).
Webster, N. S. & Taylor, M. W. Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. 14, 335–346 (2012).
Won, Y.-J., Jones, W. J. & Vrijenhoek, R. C. Absence of cospeciation between deep-sea mytilids and their thiotrophic endosymbionts. J. Shellfish Res. 27, 129–138 (2008).
Usher, K. M. & Ereskovsky, A. V. Larval development, ultrastructure and metamorphosis in Chondrilla australiensis Carter, 1873 (Demospongiae, Chondrosida, Chondrillidae). Invertebr. Reprod. Dev. 47, 51–62 (2005).
Schmitt, S., Weisz, J. B., Lindquist, N. & Hentschel, U. Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl. Environ. Microbiol. 73, 2067–2078 (2007).
Sharp, K. H., Eam, B., Faulkner, D. J. & Haygood, M. G. Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73, 622–629 (2007).
Lee, O. O., Chui, P. Y., Wong, Y. H., Pawlik, J. R. & Qian, P. Y. Evidence for vertical transmission of bacterial symbionts from adult to embryo in the Caribbean sponge Svenzea zeai. Appl. Environ. Microbiol. 75, 6147–6156 (2009).
Webster, N. S. et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12, 2070–2082 (2010). The first study to apply 16S rRNA gene pyrosequencing to sponge bacteria, revealing the high degree of overlap between bacterial communities in adult and larval sponges.
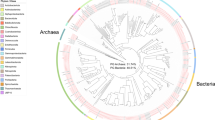
Hentschel, U. et al. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68, 4431–4440 (2002). The first molecular (16S rRNA gene) study to identify monophyletic sponge-specific clusters of bacteria.
Simister, R. L., Deines, P., Botte, E. S., Webster, N. S. & Taylor, M. W. Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ. Microbiol. 14, 517–524 (2012).
Wilkinson, C. R. Immunological evidence for the Precambrian origin of bacterial symbioses in marine sponges. Proc. R. Soc. Lond. B 220, 509–517 (1984).
Taylor, M. W., Thacker, R. W. & Hentschel, U. Genetics: evolutionary insights from sponges. Science 316, 1854–1855 (2007).
Lee, O. O. et al. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J. 5, 650–664 (2011).
Schmitt, S. et al. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 6, 564–576 (2012). An investigation that uses 16S rRNA gene pyrosequencing to describe the biogeography and host specificity of bacteria in 32 sponge species from eight worldwide locations.
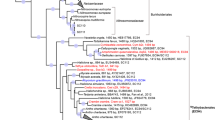
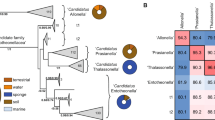
Siegl, A. et al. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 5, 61–70 (2011). The first study to apply single-cell techniques to investigate the genomes of uncultivated bacterial symbionts of sponges.
Fieseler, L., Horn, M., Wagner, M. & Hentschel, U. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70, 3724–3732 (2004).
Preston, C. M., Wu, K. Y., Molinski, T. F. & DeLong, E. F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl Acad. Sci. USA 93, 6241–6246 (1996).
Pape, T. et al. Dense populations of Archaea associated with the demosponge Tentorium semisuberites Schmidt, 1870 from Arctic deep-waters. Polar Biol. 29, 662–667 (2006).
Radax, R., Hoffmann, F., Rapp, H. T., Leininger, S. & Schleper, C. Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ. Microbiol. 14, 909–923 (2012).
Wilkinson, C. R. in Algae and Symbioses: Plants, Animals, Fungi, Viruses, Interactions Explored (ed. W. Reisser) 111–151 (Biopress Limited, 1992).
Höller, U. et al. Fungi from marine sponges: diversity, biological activity and secondary metabolites. Mycol. Res. 104, 1354–1365 (2000).
Baker, P. W., Kennedy, J., Dobson, A. D. W. & Marchesi, J. R. Phylogenetic diversity and antimicrobial activities of fungi associated with Haliclona simulans isolated from Irish coastal waters. Mar. Biotechnol. 11, 540–547 (2009).
Quince, C. et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nature Methods 6, 639–641 (2009).
Kunin, V., Engelbrektson, A., Ochman, H. & Hugenholtz, P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12, 118–123 (2010).
Sunagawa, S., Woodley, C. M. & Medina, M. Threatened corals provide underexplored microbial habitats. PLoS ONE 5, e9554 (2010).
Cardenas, A., Rodriguez, L. M., Pizarro, V., Cadavid, L. F. & Arevalo-Ferro, C. Shifts in bacterial communities of two caribbean reef-building coral species affected by white plague disease. ISME J. 123, 502–512 (2012).
Behrendt, L. et al. Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. ISME J. 6, 1222–1237 (2012).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. Microbiol. 6, 776–888 (2008).
Dewhirst, F. E. et al. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010).
Griffen, A. L. et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185 (2012).
Zoetendal, E. G., Rajilic-Stojanovic, M. & de Vos, W. M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57, 1605–1615 (2008).
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Grice, E. A. & Segre, J. A. The skin microbiome. Nature Rev. Microbiol. 9, 244–253 (2011).
Taylor, M. W., Schupp, P. J., Dahllof, I., Kjelleberg, S. & Steinberg, P. D. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6, 121–130 (2004).
Wiedenbeck, J. & Cohan, F. M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976 (2011).
Dobrindt, U., Hochhut, B., Hentschel, U. & Hacker, J. Genomic islands in pathogenic and environmental microorganisms. Nature Rev. Microbiol. 2, 414–424 (2004).
Sobecky, P. A. & Hazen, T. H. Horizontal gene transfer and mobile genetic elements in marine systems. Methods Mol. Biol. 532, 435–453 (2009).
Villareal, L. P. Viruses and the Evolution of Life (American Society for Microbiology Press, 2005).
Suttle, C. A. Marine viruses--major players in the global ecosystem. Nature Rev. Microbiol. 5, 801–812 (2007).
Claverie, J.-M. et al. Mimivirus and mimiviridae: giant viruses with an increasing number of potential hosts, including corals and sponges. J. Invertebr. Pathol. 101, 172–180 (2009).
Thomas, T. et al. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 4, 1557–1567 (2010). The first metagenomic analysis of sponge microbiota, revealing insights into mechanisms of bacterium–sponge interactions and highlighting ANK-containing proteins as putative symbiosis factors.
Grimson, A. et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455, 1193–1197 (2008).
Taylor, M. W., Hill, R. T. & Hentschel, U. Meeting report: first international symposium on sponge microbiology. Mar. Biotechnol. 13, 1057–1061 (2011).
Hallam, S. J. et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl Acad. Sci. USA 103, 18296–18301 (2006). The sequenced genome of C. symbiosum provides many new insights into the metabolic potential of this sponge symbiont.
Hallam, S. J. et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4, e95 (2006).
Liu, M. Y., Kjelleberg, S. & Thomas, T. Functional genomic analysis of an uncultured δ-proteobacterium in the sponge Cymbastela concentrica. ISME J. 5, 427–435 (2011).
Radax, R. et al. Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ. Microbiol. 14, 1308–1324 (2012).
Lopez-Legentil, S., Erwin, P. M., Pawlik, J. R. & Song, B. Effects of sponge bleaching on ammonia-oxidizing Archaea: distribution and relative expression of ammonia monooxygenase genes associated with the barrel sponge Xestospongia muta. Microb. Ecol. 60, 561–571 (2010).
Turque, A. S. et al. Environmental shaping of sponge associated archaeal communities. PLoS ONE 5, e15774 (2010).
Steger, D. et al. Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ. Microbiol. 10, 1087–1094 (2008).
Bayer, K., Schmitt, S. & Hentschel, U. Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ. Microbiol. 10, 2942–2955 (2008).
Hoffmann, F. et al. Complex nitrogen cycling in the sponge Geodia barretti. Environ. Microbiol. 11, 2228–2243 (2009).
Liu, M. Y., Fan, L., Zhong, L., Kjelleberg, S. & Thomas, T. Metaproteogenomic analysis of a community of sponge symbionts. ISME J. 6, 1515–1525 (2012). The first proteomic analysis of a sponge-associated microbiota, providing novel information on the activities, physiology and interactions of microorganisms with sponge hosts.
Gill, S. R. et al. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 (2006).
Goodman, A. L. et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 (2009).
Cazalet, C. et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nature Genet. 36, 1165–1173 (2004).
Pan, X., Lührmann, A., Satoh, A., Laskowski-Arce, M. A. & Roy, C. R. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320, 1651–1654 (2008).
Voth, D. E. ThANKs for the repeat: intracellular pathogens exploit a common eukaryotic domain. Cell. Logist. 1, 128–132 (2011).
Schmitz-Esser, S. et al. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J. Bacteriol. 192, 1045–1057 (2010).
Al-Khodor, S., Price, C. T., Kalia, A. & Abu Kwaik, Y. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 18, 132–139 (2010).
Al-Khodor, S., Price, C. T., Habyarimana, F., Kalia, A. & Abu Kwaik, Y. A. Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70, 908–923 (2008).
Price, C. T., Al-Khodor, S., Al-Quadan, T. & Abu Kwaik, Y. Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect. Immun. 78, 2079–2088 (2010).
Lührmann, A., Nogueira, C. V., Carey, K. L. & Roy, C. R. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc. Natl Acad. Sci. USA 107, 18997–19001 (2010).
D'Andrea, L. D. & Regan, L. TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662 (2003).
Mittl, P. R. & Schneider-Brachert, W. Sel1-like repeat proteins in signal transduction. Cell. Signal. 19, 20–31 (2007).
Kataeva, I. A. et al. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68, 4292–4300 (2002).
Exposito, J. Y. et al. Demosponge and sea anemone fibrillar collagen diversity reveals the early emergence of A/C clades and the maintenance of the modular structure of type V/XI collagens from sponge to human. J. Biol. Chem. 283, 28226–28235 (2008).
Har-el, R. & Tanzer, M. L. Extracellular matrix. 3: evolution of the extracellular matrix in invertebrates. FASEB J. 7, 1115–1123 (1993).
Ozbek, S., Balasubramanian, P. G., Chiquet-Ehrismann, R., Tucker, R. P. & Adams, J. C. The evolution of extracellular matrix. Mol. Biol. Cell 21, 4300–4305 (2010).
Fuqua, C. & Greenberg, E. P. Listening in on bacteria: acyl-homoserine lactone signalling. Nature Rev. Mol. Cell Biol. 3, 685–695 (2002).
Mohamed, N. M. et al. Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ. Microbiol. 10, 75–86 (2008).
Zan, J., Fricke, W. F., Fuqua, C., Ravel, J. & Hill, R. T. Genome sequence of Ruegeria sp. strain KLH11, an N-acylhomoserine lactone-producing bacterium isolated from the marine sponge Mycale laxissima. J. Bacteriol. 193, 5011–5012 (2011).
Zan, J., Fuqua, C. & Hill, R. T. Diversity and functional analysis of luxS genes in vibrios from marine sponges Mycale laxissima and Ircinia strobilina. ISME J. 5, 1505–1516 (2011). A report on quorum sensing, the presence of luxS genes and autoinducer 2 activity in sponge-associated bacterial communities.
Taylor, M. W. et al. Evidence for acyl homoserine lactone signal production in bacteria associated with marine sponges. Appl. Environ. Microbiol. 70, 4387–4389 (2004).
Hughes, D. T. & Sperandio, V. Inter-kingdom signalling: communication between bacteria and their hosts. Nature Rev. Microbiol. 6, 111–120 (2008). A summary of recently acquired evidence that bacterial quorum sensing signalling can also be used in crosstalk between microorganisms and their hosts.
Webster, N. S. Sponge disease: a global threat? Environ. Microbiol. 9, 1363–1375 (2007).
Wilson, H. V. On some phenomena of coalescence and regeneration in sponges. J. Exp. Zool. 5, 245–258 (1907).
Gordon, S. Pattern recognition receptors: doubling up for the innate immune response. Cell 111, 927–930 (2002).
Janeway, C. A. & Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 (2002).
Liu, L. et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 320, 379–381 (2008).
Dunne, A. & O'Neill, L. A. The Interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci. STKE 2003, re3 (2003).
Påisson-McDermott, E. M. & O'Neill, L. A. J. Building an immune system from nine domains. Biochem. Soc. Trans. 35, 1437–1444 (2007).
Bosch, T. C. G. et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev. Comp. Immunol. 33, 559–569 (2009).
Franchi, L., Eigenbrod, T., Munoz-Planillo, R. & Nunez, G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nature Immunol. 10, 241–247 (2009).
Elinav, E., Strowig, T., Henao-Mejia, J. & Flavell, R. A. Regulation of the antimicrobial response by NLR proteins. Immunity 34, 665–679 (2011).
Monie, T. P., C. E., B. & Gay, N. J. Activating immunity: lessons from the TLRs and NLRs. Trends Biochem. Sci. 34, 553–561 (2009).
Seong, S. & Matzinger, P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nature Rev. Immunol. 4, 469–478 (2004).
Sarrias, M. R. et al. The Scavenger Receptor Cysteine-Rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 24, 1–37 (2004).
Plüddemann, A. et al. SR-A, MARCO and TLRs differentially recognise selected surface proteins from Neisseria meningitidis: an example of fine specificity in microbial ligand recognition by innate immune receptors. Innate Immun. 1, 153–163 (2009).
Pahler, S., Blumbach, B., Muller, I. & Muller, W. E. Putative multiadhesive protein from the marine sponge Geodia cydonium: cloning of the cDNA encoding a fibronectin-, an SRCR-, and a complement control protein module. J. Exp. Zool. 282, 332–343 (1998).
Pancer, Z., Münker, J., Müller, I. M. & Müller, W. E. G. A novel member of an ancient superfamily: sponge (Geodia cydonium, Porifera) putative protein that features scavenger receptor cysteine-rich repeats. Gene 193, 211–218 (1997).
Steindler, L. et al. Differential gene expression in a marine sponge in relation to its symbiotic state. Mar. Biotechnol. 9, 543–549 (2007).
Huyck, T. K., Gradishar, W., Manuguid, F. & Kirkpatrick, P. Eribulin mesylate. Nature Rev. Drug Discov. 10, 173–174 (2011).
Aicher, T. D. et al. Total synthesis of halichondrin B and norhalichondrin B. J. Am. Chem. Soc. 114, 3162–3164 (1992).
Munro, M. G. H. et al. The discovery and development of marine compounds with pharmaceutical potential. J. Biotechnol. 70, 15–25 (1999).
Bewley, C. A. & Faulkner, D. J. Lithistid sponges: star performers or hosts to the stars. Angew. Chem. Int. Ed. Engl. 37, 2163–2178 (1998).
Faulkner, D. J., Unson, M. D. & Bewley, C. A. The chemistry of some sponges and their symbionts. Pure Appl. Chem. 66, 1983–1990 (1994).
Unson, M. D. & Faulkner, D. J. Cyanobacterial symbiont biosynthesis of chlorinated metabolites from Dysidea herbacea (Porifera). Experientia 49, 349–353 (1993). The first convincing experimental evidence that sponge-derived natural products can be of bacterial origin.
Unson, M. D., Holland, N. D. & Faulkner, D. J. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol. 119, 1–11 (1994).
Schmidt, E. W., Obraztsova, A. Y., Davidson, S. K., Faulkner, D. J. & Haygood, M. G. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis”. Mar. Biol. 136, 969–977 (2000).
Bewley, C. A., Holland, N. D. & Faulkner, D. J. Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Experientia 52, 716–722 (1996).
Gillor, O., Carmeli, S., Rahamim, Y., Fishelson, Z. & Ilan, M. Immunolocalization of the toxin latrunculin B within the Red Sea sponge Negombata magnifica (Demospongiae, Latrunculiidae). Mar. Biotechnol. 2, 213–223 (2000).
Fischbach, M. A. & Walsh, C. T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006).
Fisch, K. M. et al. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nature Chem. Biol. 5, 494–501 (2009). A report of a strategy that allows the identification of biosynthetic-gene clusters from microbial metagenomes on the basis of chemical moieties in the compounds produced by the microbial community. Using this strategy, the complete pathway for psymberin is recovered from the sponge P. bulbosa.
Piel, J. et al. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc. Natl Acad. Sci. USA 101, 16222–16227 (2004). An investigation that provides unambiguous evidence for the bacterial production of a potent antitumor compound from the sponge T. swinhoei.
Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 27, 996–1047 (2010).
Piel, J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl Acad. Sci. USA 99, 14002–14007 (2002).
Sudek, S. et al. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 70, 67–74 (2007).
Partida-Martinez, L. P. & Hertweck, C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437, 884–888 (2005).
Partida-Martinez, L. P. & Hertweck, C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chembiochem 8, 41–45 (2007).
Donia, M. S. et al. Complex microbiome underlying secondary and primary metabolism in the tunicate- Prochloron symbiosis. Proc. Natl Acad. Sci. USA 108, e1423–e1432 (2011).
Nguyen, T. et al. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nature Biotech. 26, 225–233 (2008).
Flatt, P. M. et al. Identification of the cellular site of polychlorinated peptide biosynthesis in the marine sponge Dysidea (Lamellodysidea) herbacea and symbiotic cyanobacterium Oscillatoria spongeliae by CARD-FISH analysis. Mar. Biol. 147, 761–774 (2005).
Lasken, R. S. Single-cell genomic sequencing using multiple displacement amplification. Curr. Opin. Microbiol. 10, 510–516 (2007).
Wang, D. J. & Bodovitz, S. Single cell analysis: the new frontier in 'omics'. Trends Biotechnol. 28, 281–290 (2010).
Siegl, A. & Hentschel, U. PKS and NRPS gene clusters from microbial symbiont cells of marine sponges by whole genome amplification. Environ. Microbiol. Rep. 2, 507–513 (2010).
Grindberg, R. V. et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS ONE 6, e18565 (2011).
Engene, N. et al. Underestimated biodiversity as a major explanation for the perceived rich secondary metabolite capacity of the cyanobacterial genus Lyngbya. Environ. Microbiol. 13, 1601–1610 (2011).
Intergovernmental Panel on Climate Change. Fourth Assessment Report of the Intergovernmental Panel on Climate Change (Geneva, 2007).
Lemoine, N., Buell, N., Hill, A. & Hill, M. in Porifera Research: Biodiversity, Innovation and Sustainability (eds M.R. Custódio, G. Lôbo-Hajdu, E. Hajdu, & G. Muricy) 419–425 (Série Livros, 2007).
Webster, N. S., Cobb, R. E. & Negri, A. P. Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2, 830–842 (2008).
Lopez-Legentil, S., Song, B., McMurray, S. E. & Pawlik, J. R. Bleaching and stress in coral reef ecosystems: hsp70 expression by the giant barrel sponge Xestospongia muta. Mol. Ecol. 17, 1840–1849 (2008).
Pantile, R. & Webster, N. Strict thermal threshold identified by quantitative PCR in the sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 431, 97–105 (2011).
Luter, H. M., Whalan, S. & Webster, N. S. Exploring the role of microorganisms in the disease-like syndrome affecting the sponge Ianthella basta. Appl. Environ. Microbiol. 76, 5736–5744 (2010).
Angermeier, H. et al. The pathology of sponge orange band disease affecting the Caribbean barrel sponge Xestospongia muta. FEMS Microb. Ecol. 75, 218–230 (2011).
Webster, N. S., Negri, A. P., Webb, R. I. & Hill, R. T. A spongin-boring α-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge, Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 232, 305–309 (2002).
Reiswig, H. M. Particle feeding in natural populations of three marine demosponges. Biol. Bull. 141, 568–591 (1971).
Reiswig, H. M. Bacteria as food for temperate-water marine sponges. Can. J. Zool. 53, 582–589 (1975).
Pile, A. J., Patterson, M. R. & Witman, J. D. In situ grazing on plankton <10μm by the boreal sponge Mycale lingua. Mar. Ecol. Prog. Ser. 141, 95–102 (1996).
Hadas, E., Marie, D., Shpigel, M. & Ilan, M. Virus predation by sponges is a new nutrient-flow pathway in coral reef food webs. Limnol. Oceanogr. 51, 1548–1550 (2006).
Maritz, K., Calcino, A., Fahey, B., Degnan, B. & Degnan, S. M. Remarkable consistency of larval supply in the spermcast-mating demosponge Amphimedon queenslandica (Hooper and van Soest). Open Mar. Biol. J. 4, 57–64 (2010).
Acknowledgements
The authors acknowledge the valuable contributions of all current and past members of their respective research groups. They are grateful to the participants of the First International Symposium on Sponge Microbiology (Würzburg, Germany, March 2011), for inspiring discussions and insights into many aspects of sponge microbiology. The pioneering efforts of early sponge microbiologists such as H. Reiswig, J. Vacelet and C. Wilkinson are also acknowledged. Funding for sponge and microbiology research in the authors' laboratories was provided by the German Research Foundation (DFG) grants SFB567/TPC3 and SFB630/TPA5 (to U.H.); by the DFG, the German Federal Ministry of Education and Research (BMBF), the Japan Society for the Promotion of Sciences, the German Academic Exchange Service (DAAD), the Human Frontier Science Program, the Max Planck Society (MPG) and the German Alexander von Humboldt Foundation (to J.P.); by the Australian Research Council and the University of Queensland (to S.M.D.); and by the University of Auckland (to M.W.T.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Benthic
-
Pertaining to organisms: living in or on the sea floor.
- Slime capsules
-
Extracellular masking layers that encapsulate bacterial cells, putatively shielding the cells from detection by a host (in the context of this Review, the sponge).
- Biologically active
-
Exhibiting activity in a biological assay (for example, a screen for antitumour agents).
- Aposymbiotic
-
Pertaining to an organism: lacking its symbionts.
- 16S rRNA gene library
-
A vector library created by the amplification and cloning of the 16S rRNA genes in a population. The library can be used for subsequent sequencing and phylogenetic analysis, as a method of describing the composition of a microbial community.
- Pyrosequencing
-
A next-generation sequencing method based on the detection of pyrophosphate release on nucleotide incorporation. The technique allows thousands or millions of sequences to be obtained from a given sample.
- Rare biosphere
-
The low-abundance microorganisms that are found in a microbial community.
- Operational taxonomic units
-
(OTUs). An arbitrary definition for a specific taxonomic grouping; for example, 97% 16S rRNA sequence similarity is frequently used to approximate a bacterial species.
- Phylotype
-
A sequence type; analogous to an operational taxonomic unit.
- Holobiont
-
A host animal and all of its associated microorganisms.
- CRISPRs
-
(Clustered regularly interspaced short palindromic repeats). Short repeats that are found in the genomes of many bacteria and archaea, and provide resistance to genetic elements such as viruses and plasmids.
- Heterotrophic
-
Pertaining to an organism: requiring dissolved organic matter for its carbon and energy sources.
- Autotrophic
-
Pertaining to an organism: growing on carbon dioxide as the sole carbon source.
- Ankyrin repeats
-
(ANKs). Common eukaryotic structural protein motifs that occur in functionally diverse proteins and mediate protein–protein interactions.
- Tetratrico peptide repeats
-
Common structural protein motifs that mediate protein–protein interactions and are frequently involved in the assembly of multiprotein complexes.
- Leucine-rich repeats
-
Structural protein motifs that are unusually rich in the hydrophobic amino acid leucine and are involved in the formation of protein–protein interactions.
- Quorum sensing
-
A chemical language by which bacteria communicate in and across populations through the use of small diffusible molecules. Bacterial production of quorum sensing molecules, as well as the bacterial response to these molecules, is correlated with population density.
- Lipopolysaccharide
-
A large endotoxic molecule consisting of a lipid and a polysaccharide; the major component of the outer membrane of Gram-negative bacteria. LPS induces a strong immune response in animals.
- Eumetazoans
-
The 'true' animals, as defined by having a nervous system and true tissues; this group includes cnidarians, ctenophores and bilaterians.
- Bilaterian
-
An animal that has three germ layers (endoderm, mesoderm and ectoderm) and bilateral symmetry; this group includes all animals except for sponges, ctenophores and cnidarians.
- Danger-associated molecular patterns
-
(DAMPS). Molecules that are released by stressed or damaged cells and act as endogenous signals to initiate a non-infectious repair (or inflammatory) response. Also called damage-associated molecular patterns.
- Complement system
-
The part of the vertebrate innate immune system that complements the activity of antibodies by opsonizing bacteria and inducing inflammatory responses which help to fight infection. Several complement-like components have also been identified in many invertebrates.
- Secondary metabolites
-
Metabolites that are not essential for the survival of the organism in which they are found (unlike primary metabolites, such as fatty acids, amino acids, and so on, which are essential).
- Epibionts
-
Organisms that live on the surface of another living organism.
- Polyketides
-
Secondary metabolites that are biosynthesized from short acyl-CoA units and often exhibit potent bioactivities. Examples are erythromycin and tetracycline.
- Non-ribosomal peptides
-
Peptides that are assembled by large multifunctional enzymes (termed non-ribosomal-peptide synthetases) and often contain non-proteinogenic amino acids. Many of these compounds (for example, cyclosporine and daptomycin) are of biomedical relevance.
- CARD–FISH
-
(Catalysed reporter deposition–fluorescence in situ hybridization). A technique that uses horseradish peroxidase-labelled oligonucleotide probes to obtain signals in samples with low-abundance targets. The signal is generated after incubation with fluorescently labelled tyramine, which is deposited at the labelling site by enzymatic polymerization.
Rights and permissions
About this article
Cite this article
Hentschel, U., Piel, J., Degnan, S. et al. Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10, 641–654 (2012). https://doi.org/10.1038/nrmicro2839
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2839
This article is cited by
-
Testacosides A–D, glycoglycerolipids produced by Microbacterium testaceum isolated from Tedania brasiliensis
Applied Microbiology and Biotechnology (2024)
-
Microbiome species diversity and seasonal stability of two temperate marine sponges Hymeniacidon perlevis and Suberites massa
Environmental Microbiome (2023)
-
Bacterial aerobic methane cycling by the marine sponge-associated microbiome
Microbiome (2023)
-
Profiling Prokaryotic Communities and Aaptamines of Sponge Aaptos suberitoides from Tulamben, Bali
Marine Biotechnology (2023)
-
Terpenes extracted from marine sponges with antioxidant activity: a systematic review
Natural Products and Bioprospecting (2023)