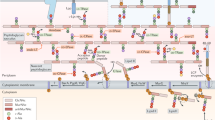
Key Points
-
Peptidoglycan synthesis is regulated at multiple levels to ensure shape-maintaining growth and cell division.
-
Peptidoglycan synthases and hydrolases coordinate to enlarge the sacculus. Coordinated enzyme activity is also required for cell division and morphogenesis.
-
Peptidoglycan synthesis and the localization and movement of cytoskeletal elements are interdependent.
-
Peptidoglycan synthases and hydrolases are also regulated by outer-membrane proteins.
-
Peptidoglycan growth is sensitive to mechanical force.
-
Peptidoglycan is remodelled in a growth-dependent manner, and its growth is tied to metabolic inputs.
Abstract
How bacteria grow and divide while retaining a defined shape is a fundamental question in microbiology, but technological advances are now driving a new understanding of how the shape-maintaining bacterial peptidoglycan sacculus grows. In this Review, we highlight the relationship between peptidoglycan synthesis complexes and cytoskeletal elements, as well as recent evidence that peptidoglycan growth is regulated from outside the sacculus in Gram-negative bacteria. We also discuss how growth of the sacculus is sensitive to mechanical force and nutritional status, and describe the roles of peptidoglycan hydrolases in generating cell shape and of D-amino acids in sacculus remodelling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Young, K. D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703 (2006).
Vollmer, W., Blanot, D. & de Pedro, M. A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 (2008).
Höltje, J.-V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62, 181–203 (1998). A landmark review on peptidoglycan synthesis in E. coli , with details of the '3 for 1' growth model.
Barreteau, H. et al. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 168–207 (2008).
Bouhss, A., Trunkfield, A. E., Bugg, T. D. & Mengin-Lecreulx, D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 32, 208–233 (2008).
Mohammadi, T. et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30, 1425–1432 (2011). An article that identifies a member of the conserved SEDS (shape, elongation, division and sporulation) family of integral membrane proteins as the elusive lipid II flippase.
Vollmer, W. & Bertsche, U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1778, 1714–1734 (2008).
Suginaka, H., Blumberg, P. M. & Strominger, J. L. Multiple penicillin-binding components in Bacillus subtilis, Bacillus cereus, Staphylococcus aureus, and Escherichia coli. J. Biol. Chem. 247, 5279–5288 (1972).
Yousif, S. Y., Broome-Smith, J. K. & Spratt, B. G. Lysis of Escherichia coli by β-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol. 131, 2839–2845 (1985).
Budd, A., Blandin, S., Levashina, E. A. & Gibson, T. J. Bacterial α2-macroglobulins: colonization factors acquired by horizontal gene transfer from the metazoan genome? Genome Biol. 5, R38 (2004).
Bertsche, U. et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 61, 675–690 (2006). The first study to provide evidence for a direct interaction between peptidoglycan synthases.
Bertsche, U., Breukink, E., Kast, T. & Vollmer, W. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. J. Biol. Chem. 280, 38096–38101 (2005).
Born, P., Breukink, E. & Vollmer, W. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. J. Biol. Chem. 281, 26985–26993 (2006). Together with reference 12, this work establishes a novel in vitro peptidoglycan assay for simultaneous detection of GTase and TPase products, demonstrating that these activities are coupled, and reconstitutes for the first time the naturally occurring reaction of TPase-mediated attachment of newly made peptidoglycan to the sacculus.
Sung, M. T. et al. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc. Natl Acad. Sci. USA 106, 8824–8829 (2009). This article describes the crystal structure of PBP1B, which includes the transmembrane region, and suggests a model for the coupling of GTase and TPase reactions.
Macheboeuf, P., Contreras-Martel, C., Job, V., Dideberg, O. & Dessen, A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30, 673–691 (2006).
Vollmer, W., Joris, B., Charlier, P. & Foster, S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 (2008).
Heidrich, C. et al. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41, 167–178 (2001).
Priyadarshini, R., Popham, D. L. & Young, K. D. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J. Bacteriol. 188, 5345–5355 (2006).
Heidrich, C., Ursinus, A., Berger, J., Schwarz, H. & Höltje, J.-V. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184, 6093–6099 (2002).
Kraft, A. R., Prabhu, J., Ursinus, A. & Höltje, J.-V. Interference with murein turnover has no effect on growth but reduces β -lactamase induction in Escherichia coli. J. Bacteriol. 181, 7192–7198 (1999).
Park, J. T. & Uehara, T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72, 211–227 (2008).
Gan, L., Chen, S. & Jensen, G. J. Molecular organization of Gram-negative peptidoglycan. Proc. Natl Acad. Sci. USA 105, 18953–18957 (2008). In this study, ECT solves a long-standing dispute about the orientation of the glycan chains in the single-layered peptidoglycan in Gram-negative bacteria.
Burman, L. G. & Park, J. T. Molecular model for elongation of the murein sacculus of Escherichia coli. Proc. Natl Acad. Sci. USA 81, 1844–1848 (1984).
Glauner, B. & Höltje, J.-V. Growth pattern of the murein sacculus of Escherichia coli. J. Biol. Chem. 265, 18988–18996 (1990). A paper that illustrates the alterations in peptidoglycan structure that occur during maturation.
Goodell, E. W., Markiewicz, Z. & Schwarz, U. Absence of oligomeric murein intermediates in Escherichia coli. J. Bacteriol. 156, 130–135 (1983).
Uehara, T. & Park, J. T. Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J. Bacteriol. 190, 3914–3922 (2008).
de Jonge, B. L. et al. Peptidoglycan synthesis during the cell cycle of Escherichia coli: composition and mode of insertion. J. Bacteriol. 171, 5783–5794 (1989).
Cabeen, M. T. & Jacobs-Wagner, C. The bacterial cytoskeleton. Annu. Rev. Genet. 44, 365–392 (2010).
Daniel, R. A. & Errington, J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113, 767–776 (2003). This work uses labelling of peptidoglycan synthesis sites to determine the topography of peptidoglycan growth in rod-shaped bacteria with or without MreB.
Jones, L. J., Carballido-Lopez, R. & Errington, J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922 (2001). A seminal paper demonstrating that MreB filaments control cell elongation.
Vats, P., Shih, Y. L. & Rothfield, L. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol. Microbiol. 72, 170–182 (2009).
Alyahya, S. A. et al. RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl Acad. Sci. USA 106, 1239–1244 (2009).
Bendezu, F. O., Hale, C. A., Bernhardt, T. G. & de Boer, P. A. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28, 193–204 (2009).
Kruse, T., Bork-Jensen, J. & Gerdes, K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55, 78–89 (2005).
Shiomi, D., Sakai, M. & Niki, H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 27, 3081–3091 (2008).
van den Ent, F. et al. Dimeric structure of the cell shape protein MreC and its functional implications. Mol. Microbiol. 62, 1631–1642 (2006).
Mohammadi, T. et al. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol. Microbiol. 65, 1106–1121 (2007).
van den Ent, F., Johnson, C. M., Persons, L., de Boer, P. & Löwe, J. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 29, 1081–1090 (2010).
Salje, J., van den Ent, F., de Boer, P. & Löwe, J. Direct membrane binding by bacterial actin MreB. Mol. Cell 43, 478–487 (2011).
Gitai, Z., Dye, N. A., Reisenauer, A., Wachi, M. & Shapiro, L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120, 329–341 (2005).
Karczmarek, A. et al. DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol. Microbiol. 65, 51–63 (2007).
Takacs, C. N. et al. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J. Bacteriol. 192, 1671–1684 (2010).
Gitai, Z., Dye, N. & Shapiro, L. An actin-like gene can determine cell polarity in bacteria. Proc. Natl Acad. Sci. USA 101, 8643–8648 (2004).
Popp, D. et al. Filament structure, organization, and dynamics in MreB sheets. J. Biol. Chem. 285, 15858–15865 (2010).
Biteen, J. S. & Moerner, W. E. Single-molecule and superresolution imaging in live bacteria cells. Cold Spring Harb. Perspect. Biol. 2, a000448 (2010).
Kim, S. Y., Gitai, Z., Kinkhabwala, A., Shapiro, L. & Moerner, W. E. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc. Natl Acad. Sci. USA 103, 10929–10934 (2006).
Dominguez-Escobar, J. et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333, 225–228 (2011).
Garner, E. C. et al. Circumferential motions of the cell wall synthesis machinery drive cytoskeletal dynamics in B. subtilis. Science 333, 222–225 (2011).
van Teeffelen, S. et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl Acad. Sci. USA 108, 15822–15827 (2011). References 47–49 describe high-resolution fluorescence microscopy showing peptidoglycan synthesis-dependent movement of MreB perpendicular to the long axis.
Kawai, Y., Daniel, R. A. & Errington, J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol. Microbiol. 71, 1131–1144 (2009).
White, C. L., Kitich, A. & Gober, J. W. Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol. Microbiol. 76, 616–633 (2010).
Land, A. D. & Winkler, M. E. Requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J. Bacteriol. 193, 4166–4179 (2011).
Adams, D. W. & Errington, J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nature Rev. Microbiol. 7, 642–653 (2009).
Erickson, H. P., Anderson, D. E. & Osawa, M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 (2010).
Aarsman, M. E. et al. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55, 1631–1645 (2005).
Aaron, M. et al. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 64, 938–952 (2007). This investigation demonstrates the FtsZ-dependent preseptal phase of cell elongation in C. crescentus.
de Pedro, M. A., Quintela, J. C., Höltje, J.-V. & Schwarz, H. Murein segregation in Escherichia coli. J. Bacteriol. 179, 2823–2834 (1997).
Fraipont, C. et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–259 (2011).
Buddelmeijer, N. & Beckwith, J. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52, 1315–1327 (2004).
Masson, S. et al. Central domain of DivIB caps the C-terminal regions of the FtsL/DivIC coiled-coil rod. J. Biol. Chem. 284, 27687–27700 (2009).
Wissel, M. C. & Weiss, D. S. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J. Bacteriol. 186, 490–502 (2004).
Rico, A. I., Garcia-Ovalle, M., Palacios, P., Casanova, M. & Vicente, M. Role of Escherichia coli FtsN protein in the assembly and stability of the cell division ring. Mol. Microbiol. 76, 760–771 (2010).
Bernard, C. S., Sadasivam, M., Shiomi, D. & Margolin, W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 64, 1289–1305 (2007).
Gerding, M. A. et al. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 191, 7383–7401 (2009).
Ursinus, A. et al. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J. Bacteriol. 186, 6728–6737 (2004).
Müller, P. et al. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J. Biol. Chem. 282, 36394–36402 (2007).
Goley, E. D. et al. Assembly of the Caulobacter cell division machine. Mol. Microbiol. 80, 1680–1698 (2011).
Briegel, A. et al. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol. Microbiol. 62, 5–14 (2006).
Li, Z. & Jensen, G. J. Electron. cryotomography: a new view into microbial ultrastructure. Curr. Opin. Microbiol. 12, 333–340 (2009).
Charbon, G., Cabeen, M. T. & Jacobs-Wagner, C. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 23, 1131–1144 (2009).
Ausmees, N., Kuhn, J. R. & Jacobs-Wagner, C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115, 705–713 (2003). This work identifies the first bacterial IF protein, CreS, which is required for the bent cell shape of C. crescentus.
Cabeen, M. T. et al. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 28, 1208–1219 (2009). A study which shows that mechanical force on the cell envelope, generated by CreS, causes C. crescentus and E. coli cells to grow with a bent shape.
Bagchi, S., Tomenius, H., Belova, L. M. & Ausmees, N. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol. Microbiol. 70, 1037–1050 (2008).
Kühn, J. et al. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 29, 327–339 (2010). An article describing the identification of a new class of bacterial cytoskeleton proteins: the bactofilins.
Koch, M. K., McHugh, C. A. & Hoiczyk, E. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol. Microbiol. 80, 1031–1051 (2011).
Sycuro, L. K. et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell 141, 822–833 (2010). This investigation demonstrates that peptidoglycan endopeptidases and a bactofilin participate in generating the helical cell shape in H. pylori.
Wang, S., Arellano-Santoyo, H., Combs, P. A. & Shaevitz, J. W. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc. Natl Acad. Sci. USA 107, 9182–9185 (2010).
Takeuchi, S., DiLuzio, W. R., Weibel, D. B. & Whitesides, G. M. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 5, 1819–1823 (2005).
Hamant, O. & Traas, J. The mechanics behind plant development. New Phytol. 185, 369–385 (2010).
Sliusarenko, O., Cabeen, M. T., Wolgemuth, C. W., Jacobs-Wagner, C. & Emonet, T. Processivity of peptidoglycan synthesis provides a built-in mechanism for the robustness of straight-rod cell morphology. Proc. Natl Acad. Sci. USA 107, 10086–10091 (2010).
Furchtgott, L., Wingreen, N. S. & Huang, K. C. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Mol. Microbiol. 81, 340–353 (2011).
Osawa, M., Anderson, D. E. & Erickson, H. P. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 28, 3476–3484 (2009).
Weart, R. B. et al. A metabolic sensor governing cell size in bacteria. Cell 130, 335–347 (2007).
Foulquier, E., Pompeo, F., Bernadac, A., Espinosa, L. & Galinier, A. The YvcK protein is required for morphogenesis via localization of PBP1 under gluconeogenic growth conditions in Bacillus subtilis. Mol. Microbiol. 80, 309–318 (2011).
Chaudhuri, R. R. et al. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH). BMC Genomics 10, 291 (2009).
Ingerson-Mahar, M., Briegel, A., Werner, J. N., Jensen, G. J. & Gitai, Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nature Cell Biol. 12, 739–746 (2010).
de Pedro, M. A., Young, K. D., Höltje, J.-V. & Schwarz, H. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185, 1147–1152 (2003).
Nelson, D. E. & Young, K. D. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182, 1714–1721 (2000).
Potluri, L. et al. Septal and lateral wall localization of PBP5, the major D,D-carboxypeptidase of Escherichia coli, requires substrate recognition and membrane attachment. Mol. Microbiol. 77, 300–323 (2010).
Markiewicz, Z., Glauner, B. & Schwarz, U. Murein structure and lack of DD- and LD-carboxypeptidase activities in Caulobacter crescentus. J. Bacteriol. 156, 649–655 (1983).
Bernhardt, T. G. & de Boer, P. A. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48, 1171–1182 (2003).
Uehara, T., Dinh, T. & Bernhardt, T. G. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol. 191, 5094–5107 (2009).
Uehara, T., Parzych, K. R., Dinh, T. & Bernhardt, T. G. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29, 1412–1422 (2010).
Peters, N. T., Dinh, T. & Bernhardt, T. G. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J. Bacteriol. 193, 4973–4983 (2011).
Goley, E. D., Comolli, L. R., Fero, K. E., Downing, K. H. & Shapiro, L. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter. Mol. Microbiol. 77, 56–73 (2010).
Möll, A., Schlimpert, S., Briegel, A., Jensen, G. J. & Thanbichler, M. DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Mol. Microbiol. 77, 90–107 (2010).
Poggio, S., Takacs, C. N., Vollmer, W. & Jacobs-Wagner, C. A protein critical for cell constriction in the Gram-negative bacterium Caulobacter crescentus localizes at the division site through its peptidoglycan-binding LysM domains. Mol. Microbiol. 77, 74–89 (2010).
Bonis, M., Ecobichon, C., Guadagnini, S., Prevost, M. C. & Boneca, I. G. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol. Microbiol. 78, 809–819 (2010).
Legaree, B. A. & Clarke, A. J. Interaction of penicillin-binding protein 2 with soluble lytic transglycosylase B1 in Pseudomonas aeruginosa. J. Bacteriol. 190, 6922–6926 (2008).
Morlot, C., Uehara, T., Marquis, K. A., Bernhardt, T. G. & Rudner, D. Z. A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis. Genes Dev. 24, 411–422 (2010). This and reference 93 show for the first time that septum-splitting peptidoglycan hydrolases require activation by other proteins.
Paradis-Bleau, C. et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143, 1110–1120 (2010).
Typas, A. et al. Regulation of peptidoglycan synthesis by outer membrane proteins. Cell 143, 1097–1109 (2010). Together with reference 101, this work demonstrates that peptidoglycan synthesis is controlled from outside the sacculus by newly identified outer-membrane lipoproteins.
Clarke, C. A., Scheurwater, E. M. & Clarke, A. J. The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J. Biol. Chem. 285, 14843–14847 (2010).
Russell, A. B. et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 (2011).
Jensen, L. J. et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 (2009).
Han, S. et al. Distinctive attributes of β-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics. J. Am. Chem. Soc. 133, 20536–20545 (2011).
Gerding, M. A., Ogata, Y., Pecora, N. D., Niki, H. & de Boer, P. A. The trans-envelope Tol–Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 63, 1008–1025 (2007).
Yeh, Y. C., Comolli, L. R., Downing, K. H., Shapiro, L. & McAdams, H. H. The Caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J. Bacteriol. 192, 4847–4858 (2010).
Zimmermann, U. Physics of turgor- and osmoregulation. Annu. Rev. Plant Physiol. 29, 121–148 (1978).
Cayley, D. S., Guttman, H. J. & Record, M. T. Jr. Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys. J. 78, 1748–1764 (2000).
Doyle, R. J. & Marquis, R. E. Elastic, flexible peptidoglycan and bacterial cell wall properties. Trends Microbiol. 2, 57–60 (1994).
Koch, A. L. Shrinkage of growing Escherichia coli cells by osmotic challenge. J. Bacteriol. 159, 919–924 (1984).
Koch, A. L. & Woeste, S. Elasticity of the sacculus of Escherichia coli. J. Bacteriol. 174, 4811–4819 (1992).
Yao, X., Jericho, M., Pink, D. & Beveridge, T. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol. 181, 6865–6875 (1999).
Sochacki, K. A., Shkel, I. A., Record, M. T. & Weisshaar, J. C. Protein diffusion in the periplasm of E. coli under osmotic stress. Biophys. J. 100, 22–31 (2011).
Vollmer, W. & Seligman, S. J. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 18, 59–66 (2010).
Jiang, H. & Sun, S. X. Morphology, growth, and size limit of bacterial cells. Phys. Rev. Lett. 105, 028101 (2010).
Glauner, B., Höltje, J.-V. & Schwarz, U. The composition of the murein of Escherichia coli. J. Biol. Chem. 263, 10088–10095 (1988).
Vollmer, W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 32, 287–306 (2008).
Lam, H. et al. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555 (2009).
Cava, F., de Pedro, M. A., Lam, H., Davis, B. M. & Waldor, M. K. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 30, 3442–3453 (2011). Together with reference 120, this paper shows that unusual D -amino acids are secreted and linked to peptidoglycan in many bacteria.
Lupoli, T. J. et al. Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc. 133, 10748–10751 (2011).
Shah, I. M., Laaberki, M. H., Popham, D. L. & Dworkin, J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135, 486–496 (2008).
Kolodkin-Gal, I. et al. D-amino acids trigger biofilm disassembly. Science 328, 627–629 (2010).
Butland, G. et al. eSGA: E. coli synthetic genetic array analysis. Nature Methods 5, 789–795 (2008).
Typas, A. et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nature Methods 5, 781–787 (2008).
Nichols, R. J. et al. Phenotypic landscape of a bacterial cell. Cell 144, 143–156 (2011). This article describes a high-throughput chemical genomic screen that provides links for the function of numerous orphan proteins in E. coli.
Andre, G. et al. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nature Commun. 1, 27 (2010).
Scheuring, S. & Dufrene, Y. F. Atomic force microscopy: probing the spatial organization, interactions and elasticity of microbial cell envelopes at molecular resolution. Mol. Microbiol. 75, 1327–1336 (2010).
Hayhurst, E. J., Kailas, L., Hobbs, J. K. & Foster, S. J. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl Acad. Sci. USA 105, 14603–14608 (2008).
Turner, R. D. et al. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nature Commun. 1, 26 (2010).
Mingorance, J. et al. Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J. Biol. Chem. 280, 20909–20914 (2005).
Huang, K. C., Mukhopadhyay, R., Wen, B., Gitai, Z. & Wingreen, N. S. Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl Acad. Sci. USA 105, 19282–19287 (2008).
Kern, T. et al. Dynamics characterization of fully hydrated bacterial cell walls by solid-state NMR: evidence for cooperative binding of metal ions. J. Am. Chem. Soc. 132, 10911–10919 (2010).
Kern, T. et al. Toward the characterization of peptidoglycan structure and protein–peptidoglycan interactions by solid-state NMR spectroscopy. J. Am. Chem. Soc. 130, 5618–5619 (2008).
Alexeeva, S., Gadella, T. W. Jr, Verheul, J., Verhoeven, G. S. & den Blaauwen, T. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol. Microbiol. 77, 384–398 (2010).
Osawa, M., Anderson, D. E. & Erickson, H. P. Reconstitution of contractile FtsZ rings in liposomes. Science 320, 792–794 (2008).
Matias, V. R., Al-Amoudi, A., Dubochet, J. & Beveridge, T. J. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 185, 6112–6118 (2003). Together with reference 82, this study demonstrates that membrane-attached FtsZ is sufficient to produce a constrictive force in vitro.
Acknowledgements
This work was supported by grants from the UK Biotechnology and Biological Sciences Research Council (BB/G015902/1 and BBI020012/1 to W.V.), the European Commission (DIVINOCELL HEALTH-F3-2009-223,431 to W.V.), the Royal Society (to W.V.) and the US National Institutes of Health (R01 GM085697, ARRA GM085697-01S1 and R01 GM036278 to C.A.G., and K99GM092984 to A.T.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Glossary
- Sacculus
-
A bag-like macromolecule that is made of peptidoglycan chains crosslinked by short peptides. The sacculus completely encases the cytoplasmic membrane in most bacteria, and isolated sacculi retain the shape of the bacterial cell.
- Bacterial cytoskeleton
-
A filamentous and often dynamic cytoplasmic structure that includes bacterial structural homologues of actin, tubulin or intermediate filaments and is essential for bacterial growth, motility, cell division, morphology and DNA segregation.
- Actin
-
A eukaryotic cytoskeletal protein with ATPase activity. MreB and ParM, two bacterial proteins involved in cell elongation and plasmid partitioning, respectively, are distant actin homologues.
- Tubulin
-
A cytoskeletal protein that forms microtubules in eukaryotes; the bacterial tubulin-like protein, FtsZ, is a GTPase and forms dynamic filaments to drive cell division.
- Penicillin-binding proteins
-
(PBPs). A protein family involved in the synthesis (the class A and class B PBPs) or hydrolysis (the class C PBPs) of D-amino acid–D-amino acid peptide bonds. They contain an active-site Ser residue that participates in the transfer of an acyl compound to an amino group or water. PBPs are the targets of β-lactam antibiotics (such as penicillin). Pathogen resistance to β-lactams can be caused by low-affinity PBPs.
- β-lactam antibiotics
-
An important class of antibiotics, members of which contain a β-lactam ring and inhibit peptidoglycan synthesis by covalent binding to the active-site Ser of penicillin-binding proteins.
- Autolysins
-
Proteins that are located in the periplasm of Gram-negative bacteria or in the cell wall of Gram-positive bacteria and can lyse the cell using their peptidoglycan-hydrolysing activity. Autolysins can have muramidase, glucosaminidase, amidase and/or endopeptidase activity.
- Electron cryotomography
-
(ECT). An electron microscopy technique that provides high-resolution pictures of an object from different angles, permitting its three-dimensional reconstitution; plunge-freezing of the samples prevents staining and fixation artefacts. In the case of the bacterial sacculus, ECT has yielded a nanometre-scale three-dimensional representation of the fine structure.
- Intermediate filaments
-
Filaments formed by coiled-coil-rich cytoskeletal proteins, such as keratin. Crescentin is a bacterial version of an intermediate filament and is required for the bent cell shape of Caulobacter crescentus.
- Blebbing
-
The release of vesicles from the outer membrane of Gram-negative bacteria. Blebbing occurs during normal growth and is enhanced in certain mutants that are impaired in cell division.
- Lysozyme
-
An antibacterial enzyme that is produced in animals, plants, fungi and even bacteria and is capable of lysing sensitive bacteria by hydrolysing the N-acetylmuramic acid–N-acetylglucosamine bonds in peptidoglycan chains.
- Type VI secretion systems
-
(T6SSs). A recently discovered secretion apparatus that is widely distributed in Gram-negative bacteria. Some of its components are similar to phage injection systems. The T6SS punctures both eukaryotic and bacterial cells, often injecting toxic effector proteins into them.
- Turgor
-
The osmotic pressure of a compartment (here, the bacterial cytoplasm) that is due to the lower activity of water.
- D-amino acids
-
Rare chiral forms (mirror structures) of the abundant L-amino acids that build proteins. D-amino acids are present in peptidoglycan and in some non-ribosomally synthesized antibiotics.
- Atomic force microscopy
-
(AFM). A microscopy technique that uses a cantilever tip to scan the surface of a probe, either in direct contact or in oscillation mode, to produce topography images with nanometre-scale resolution.
- Total internal reflection fluorescence microscopy
-
(TIRF microscopy). A fluorescence microscopy technique that uses an evanescent wave to selectively excite a fluorophore in a small area of a specimen adjacent to a glass–water interface to reduce background fluorescence. This technique provides a superior axial resolution.
- Photoactivated localization microscopy
-
(PALM). A super-resolution fluorescence microscopy technique based on the controlled activation and sampling of subsets of photoconvertible fluorescent molecules in the sample. This technique can achieve 10–20 nm resolution.
- Solid-state NMR spectroscopy
-
NMR spectroscopy of insoluble polymers. The technique requires rapid spinning of the sample at a certain 'magic' angle. It provides information on the structural flexibility of a polymer and the interactions of chemical entities within it (for example, amino acids or sugars in peptidoglycan sacculi).
- Förster resonance energy transfer
-
(FRET). A technique that detects and characterizes the interaction between two molecules coupled to two fluorophores, by measuring the excitation of one fluorophore by the light emitted from the other. A positive FRET signal indicates a distance of less than 10 nm between the fluorophores.
Rights and permissions
About this article
Cite this article
Typas, A., Banzhaf, M., Gross, C. et al. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10, 123–136 (2012). https://doi.org/10.1038/nrmicro2677
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2677
This article is cited by
-
Bacillus cereus extracellular vesicles act as shuttles for biologically active multicomponent enterotoxins
Cell Communication and Signaling (2023)
-
Exploring the potential of Rhizopus oryzae AUMC14899 as a novel endophytic fungus for the production of l-tyrosine and its biomedical applications
Microbial Cell Factories (2023)
-
Bacteria evolve macroscopic multicellularity by the genetic assimilation of phenotypically plastic cell clustering
Nature Communications (2023)
-
Multicopy suppressor screens reveal convergent evolution of single-gene lysis proteins
Nature Chemical Biology (2023)
-
Two broadly conserved families of polyprenyl-phosphate transporters
Nature (2023)