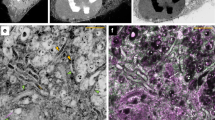
Abstract
Cryo-electron tomography is an emerging imaging technique that has unique potential for molecular cell biology. At the present resolution of 4–5 nm, large supramolecular structures can be studied in unperturbed cellular environments and, in the future, it will become possible to map molecular landscapes inside cells in a more comprehensive manner. 'Visual proteomics' aims to complement and extend mass-spectrometry-based inventories, and to provide a quantitative description of the macromolecular interactions that underlie cellular functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Frey-Wyssling, A. The submicroscopic structure of the cytoplasm. J. R. Microsc. Soc. 60, 128–139 (1940).
Frey-Wyssling, A. Submicroscopic Morphology of Protoplasm and its Derivatives (Elsevier, New York, 1948).
Afzelius, B. A. & Maunsbach, A. B. Biological ultrastructure research; the first 50 years. Tissue Cell 36, 83–94 (2004).
Ball, P. Portrait of a molecule. Nature 421, 421–422 (2003).
Baumeister, W. From proteomic inventory to architecture. FEBS Lett. 579, 933–937 (2005).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Huh, W.-K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Phizicky, E., Bastiaens, P. I. H., Zhu, H., Snyder, M. & Fields, S. Protein analysis on a proteomic scale. Nature 422, 208–215 (2003).
Hell, S. W., Dyba, M. & Jakobs, S. Concepts for nanoscale resolution in fluorescence microscopy. Curr. Opin. Neurobiol. 14, 599–609 (2004).
Miyawaki, A., Sawano, A. & Kogure, T. Lighting up cells: labelling proteins with fluorophores. Nature Cell Biol. 5, S1–S7 (2003).
Grunewald, K., Medalia, O., Gross, A., Steven, A. C. & Baumeister, W. Prospects of electron cryotomography to visualize macromolecular complexes inside cellular compartments: implications of crowding. Biophys. Chem. 100, 577–591 (2003).
Koster, A. J. et al. Perspectives of molecular and cellular electron tomography. J. Struct. Biol. 120, 276–308 (1997).
Grimm, R. et al. Energy filtered electron tomography of ice-embedded actin and vesicles. Biophys. J. 72, 482–489 (1997).
Grimm, R. et al. Electron tomography of ice-embedded prokaryotic cells. Biophys. J. 74, 1031–1042 (1998).
Dierksen, K. et al. Three-dimensional structure of lipid vesicles embedded in vitreous ice and investigated by automated electron tomography. Biophys. J. 68, 1416–1422 (1995).
Wolosewick, J. J. & Porter, K. R. Stereo high-voltage electron-microscopy of whole cells of human diploid line, WI-38. Am. J. Anat. 147, 303–323 (1976).
Heuser, J. Whatever happened to the 'microtrabecular concept'? Biol. Cell 94, 561–596 (2003).
Matricardi, V. R., Moretz, R. C. & Parsons, D. F. Electron-diffraction of wet proteins — catalase. Science 177, 268–270 (1972).
Parsons, D. F. Structure of wet specimens in electron-microscopy. Science 186, 407–414 (1974).
Taylor, K. A. & Glaeser, R. M. Electron-diffraction of frozen, hydrated protein crystals. Science 186, 1036–1037 (1974).
Burton, E. F. & Oliver, W. F. The crystal structure of ice at low temperature. Proc. R. Soc. Lond. A 153, 166–172 (1935).
Mayer, E. & Brüggeller, P. Complete vitrification in pure liquid water and dilute aqueous solutions. Nature 288, 569–571 (1980).
Dubochet, J. & McDowall, A. W. Vitrification of pure water for electron microscopy. J. Microsc. 124, RP3–RP4 (1981).
Steven, A. C. & Aebi, U. The next ice age: cryo-electron tomography of intact cells. Trends Cell Biol. 13, 107–110 (2003).
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988).
McIntosh, J. R., Nicastro, D. & Mastronarde, D. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 15, 43–51 (2005).
Marsh, B. J. Lessons from tomographic studies of the mammalian Golgi. Biochim. Biophys. Acta 1744, 273–292 (2005).
Nicastro, D., McIntosh, J. R. & Baumeister, W. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl Acad. Sci. USA 102, 15889–15894 (2005).
Zhang, P. et al. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J. Microsc. 216, 76–83 (2004).
Al-Amoudi, A. et al. Cryo-electron microscopy of vitreous sections. EMBO J. 23, 3583–3588 (2004).
Medalia, O. et al. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 298, 1209–1213 (2002).
Grunewald, K. et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302, 1396–1398 (2003).
Beck, M. et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306, 1387–1390 (2004).
Mercogliano, C. P. & DeRosier, D. J. Gold nanocluster formation using metallothionein: mass spectrometry and electron microscopy. J. Mol. Biol. 355, 211–223 (2006).
Bohm, J. et al. Toward detecting and identifying macromolecules in a cellular context: template matching applied to electron tomograms. Proc. Natl Acad. Sci. USA 97, 14245–14250 (2000).
Frangakis, A. S. et al. Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc. Natl Acad. Sci. USA 99, 14153–14158 (2002).
Frangakis, A. S. & Forster, F. Computational exploration of structural information from cryo-electron tomograms. Curr. Opin. Struct. Biol. 14, 325–331 (2004).
Sali, A., Glaeser, R., Earnest, T. & Baumeister, W. From words to literature in structural proteomics. Nature 422, 216–225 (2003).
Aloy, P. & Russell, R. B. Structure-based systems biology: a zoom lens for the cell. FEBS Lett. 579, 1854–1858 (2005).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Glaeser, R. M. Historical background: why is it important to improve automated particle selection methods? J. Struct. Biol. 145, 15–18 (2004).
Radon, J. Über die Bestimmung von Funktionen durch ihre Integralwerte längs gewisser Mannigfaltigkeiten. Berichte über die Verhandlungen der Königlich Sächsischen Gesellschaft der Wissenschaften zu Leipzig. Math. Phys. Klasse 69, 262–277 (1917) (in German).
Bracewell, R. N. & Riddle, A. C. Inversion of fan-beam scans in radio astronomy. Astrophys. J. 150, 427–434 (1967).
DeRosier, D. J. & Klug, A. Reconstruction of three dimensional structures from electron micrographs. Nature 217, 130–134 (1968).
Unwin, P. N. T. & Henderson, R. Molecular-structure determination by electron-microscopy of unstained crystalline specimens. J. Mol. Biol. 94, 425–440 (1975).
Henderson, R. & Unwin, P. N. T. Three-dimensional model of purple membrane obtained by electron-microscopy. Nature 257, 28–32 (1975).
Frank, J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu. Rev. Biophys. Biomol. Struct. 31, 303–319 (2002).
Hart, R. G. Electron microscopy of unstained material: the polytropic montage. Science 159, 1464–1467 (1968).
Lücić, V., Foerster, F. & Baumeister, W. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biophys. Biomol. Struct. 74, 833–865 (2005).
Leis, A. et al. Cryo-electron tomography and fluorescence microscopy of unicellular algae in vitreous cryosections. Microsc. Microanal. 11 (Suppl. 2), 330–331 (2005).
Ruepp, A. et al. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407, 508–513 (2000).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
FURTHER INFORMATION
3D electron microscopy at Utrecht University
Cryo-electron tomography of Dictyostelium discoideum (movie)
High Resolution Electron Microscopy at the National Cancer Institute
Institute for Molecular Bioscience, The University of Queensland, Australia
Max-Planck-Institute for Biochemistry, Department of Molecular Structural Biology
Molecular Art | Molecular Science, Home of David S. Goodsell
Resource for the Visualization of Biological Complexity, Wadsworth Center
The Boulder Laboratory for 3-D Electron Microscopy of Cells
The Jensen Laboratory for Cryo-Electron Microscopy
The Three Dimensional Electron Microscopy (3D-EM) Network of Excellence
University of California San Francisco, Macromolecular Structure Group
Rights and permissions
About this article
Cite this article
Nickell, S., Kofler, C., Leis, A. et al. A visual approach to proteomics. Nat Rev Mol Cell Biol 7, 225–230 (2006). https://doi.org/10.1038/nrm1861
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm1861
This article is cited by
-
Mysterious Asgard archaea microbes reveal their inner secrets
Nature (2023)
-
Searching for 3D structural models from a library of biological shapes using a few 2D experimental images
BMC Bioinformatics (2018)
-
Improved deep learning-based macromolecules structure classification from electron cryo-tomograms
Machine Vision and Applications (2018)
-
Simulating cryo electron tomograms of crowded cell cytoplasm for assessment of automated particle picking
BMC Bioinformatics (2016)
-
Using electron microscopes to look into the lung
Histochemistry and Cell Biology (2016)