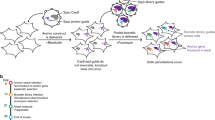
Key Points
-
Functional annotation of human genes has been hampered by the lack of tools to probe gene function systematically and quickly. RNA interference (RNAi) offers the cell biologist an approach to perturb gene function that can be applied in a high-throughput fashion on the cell or organism scale.
-
Various approaches to RNAi-mediated gene knockdown have been engineered for screening gene function in mammalian cells, including small interfering RNAs (siRNAs), plasmid-based short hairpin RNAs (shRNAs), plasmid-based shRNAs in a microRNA context (shRNA-mirs), virus-based shRNAs (retroviral and lentiviral), enzymatically prepared siRNAs (esiRNAs) and others. This review surveys several of the screens that were undertaken using these approaches to show their applicability.
-
A number of different formats for high-throughput mammalian RNAi screens have been developed and are discussed, including well-based arrayed screening, pooled screening and cell microarrays.
-
Methods for undertaking a mammalian RNAi screen focus on component discovery, component validation, component classification and systems-type analysis. Ideas for navigating through each of these screening steps are discussed.
-
A road map for RNAi screening using a hypothetical example to look for regulators of cell growth and proliferation is presented as an example of how the architecture of a signalling pathway could quickly be constructed using RNAi gene-knockdown technology.
-
Further development of RNAi technology will provide greater control over gene expression as well as advancing organ and organismal studies.
Abstract
Technological advances in mammalian systems are providing new tools to identify the molecular components of signalling pathways. Foremost among these tools is the ability to knock down gene function through the use of RNA interference (RNAi). The fact that RNAi can be scaled up for use in high-throughput techniques has motivated the creation of genome-wide RNAi reagents. We are now at the brink of being able to harness the power of RNAi for large-scale functional discovery in mammalian cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brummelkamp, T. R. & Bernards, R. New tools for functional mammalian cancer genetics. Nature Rev. Cancer 3, 781?789 (2003).
Hannon, G. J. & Rossi, J. J. Unlocking the potential of the human genome with RNA interference. Nature 431, 371?378 (2004).
Sharp, P. A. RNAi and double-strand RNA. Genes Dev. 13, 139?141 (1999).
Novina, C. D. & Sharp, P. A. The RNAi revolution. Nature 430, 161?164 (2004).
Meister, G. & Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343?349 (2004).
Zamore, P. D. & Haley, B. Ribo-gnome: the big world of small RNAs. Science 309, 1519?1524 (2005).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806?811 (1998). First demonstration in animals that dsRNA can target gene-specific knockdown.
Fraser, A. G. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325?330 (2000).
Kamath, R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231?237 (2003).
Ashrafi, K. et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268?272 (2003).
Lum, L. et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299, 2039?2045 (2003).
Boutros, M. et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832?835 (2004).
Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. How cells respond to interferons. Annu. Rev. Biochem. 67, 227?264 (1998).
Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494?498 (2001). Seminal discovery that short dsRNAs trigger gene silencing in mammalian cells in the absence of nonspecific responses.
McManus, M. T., Petersen, C. P., Haines, B. B., Chen, J. & Sharp, P. A. Gene silencing using micro-RNA designed hairpins. RNA 8, 842?850 (2002).
Brummelkamp, T. R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550?553 (2002). One of the first demonstrations that plasmid-based shRNAs can target gene-specific knockdown.
Abbas-Terki, T., Blanco-Bose, W., Deglon, N., Pralong, W. & Aebischer, P. Lentiviral-mediated RNA interference. Hum. Gene Ther. 13, 2197?2201 (2002).
Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16, 948?958 (2002).
Stewart, S. A. et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493?501 (2003).
Cullen, B. R. Transcription and processing of human microRNA precursors. Mol. Cell 16, 861?865 (2004).
Zeng, Y., Wagner, E. J. & Cullen, B. R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9, 1327?1333 (2002).
Silva, J. M. et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature Genet. 37, 1281?1288 (2005). Describes the first shRNA-mir library that targets all human and mouse genes with 2?3 constructs per gene.
Manche, L., Green, S. R., Schmedt, C. & Mathews, M. B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12, 5238?5248 (1992).
Minks, M. A., West, D. K., Benvin, S. & Baglioni, C. Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J. Biol. Chem. 254, 10180?10183 (1979).
Berns, K. et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428, 431?437 (2004). The first large-scale retroviral library that was used in a pooled infection screen.
Paddison, P. J. et al. A resource for large-scale RNA-interference-based screens in mammals. Nature 428, 427?431 (2004).
Arts, G. J. et al. Adenoviral vectors expressing siRNAs for discovery and validation of gene function. Genome Res. 13, 2325?2332 (2003).
Moffat, J. et al. A lentiviral RNAi library targeting human and mouse genes: construction, characterization and application to an arrayed high-content screen. Cell (in the press). Describes the first large-scale lentiviral shRNA library and arrayed viral infection screen.
Aza-Blanc, P. et al. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell 12, 627?637 (2003).
Mackeigan, J. P., Murphy, L. O. & Blenis, J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nature Cell Biol. 7, 591?600 (2005).
Pelkmans, L et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436, 78?86 (2005). Describes the first genome-wide siRNA transfection screen of human kinases that are required for endocytosis and a systems-level analysis of the screening results.
Ovcharenko, D., Jarvis, R., Hunicke-Smith, S., Kelnar, K. & Brown, D. High-throughput RNAi screening in vitro: from cell lines to primary cells. RNA 11, 985?993 (2005).
Brummelkamp, T. R., Nijman, S. M., Dirac, A. M. & Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424, 797?801 (2003).
Cai, X., Hagedorn, C. H. & Cullen, B. R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957?1966 (2004).
Lee, Y. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051?4060 (2004).
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415?419 (2003).
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231?235 (2004).
Landthaler, M., Yalcin, A. & Tuschl, T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14, 2162?2167 (2004).
Han, J. et al. The Drosha?DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016?3027 (2004).
Gregory, R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235?240 (2004).
Yi, R., Qin, Y., Macara, I. G. & Cullen, B. R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011?3016 (2003).
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E. & Kutay, U. Nuclear export of microRNA precursors. Science 303, 95?98 (2004).
Dickins, R. A. et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature Genet. 37, 1289?1295 (2005). Elegant study that demonstrates the potential of shRNA-mirs in vitro and in vivo.
Stegmeier, F., Hu, G., Rickles, R. J., Hannon, G. J. & Elledge, S. J. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA 102, 13212?13217 (2005).
Paddison, P. J., Caudy, A. A. & Hannon, G. J. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl Acad. Sci. USA 99, 1443?1448 (2002).
Sen, G., Wehrman, T. S., Myers, J. W. & Blau, H. M. Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nature Genet. 36, 183?189 (2004).
Shirane, D. et al. Enzymatic production of RNAi libraries from cDNAs. Nature Genet. 36, 190?196 (2004).
Luo, B., Heard, A. D. & Lodish, H. F. Small interfering RNA production by enzymatic engineering of DNA (SPEED). Proc. Natl Acad. Sci. USA 101, 5494?5499 (2004).
Kittler, R. et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432, 1036?1040 (2004).
Zheng, L. et al. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc. Natl Acad. Sci. USA 101, 135?140 (2004).
Sandy, P., Ventura, A. & Jacks, T. Mammalian RNAi: a practical guide. Biotechniques 39, 215?224 (2005).
Downward, J. Use of RNA interference libraries to investigate oncogenic signalling in mammalian cells. Oncogene 23, 8376?8383 (2004).
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901?906 (1999).
Westbrook, T. F. et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 121, 837?848 (2005).
Kolfschoten, I. G. et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell 121, 849?858 (2005).
Wheeler, D. B., Carpenter, A. E. & Sabatini, D. M. Cell microarrays and RNA interference chip away at gene function. Nature Genet. 37, S25?S30 (2005).
Bailey, S. N., Ali, S. M., Carpenter, A. E., Higgins, C. O. & Sabatini, D. M. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nature Meth. 3, 117?122 (2006).
Bettencourt-Dias, M. et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432, 980?987 (2004).
Zhang, J. H., Chung, T. D. & Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67?73 (1999).
Bridge, A. J., Pebernard, S., Ducraux, A., Nicoulaz, A. L. & Iggo, R. Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genet. 34, 263?264 (2003).
Jackson, A. L. & Linsley, P. S. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 20, 521?524 (2004).
Hoffmann, R. & Valencia, A. A gene network for navigating the literature. Nature Genet. 36, 664 (2004).
Hoffmann, R. et al. Text mining for metabolic pathways, signaling cascades, and protein networks. Sci. STKE 283, pe21 (2005).
Hoffmann, R. & Valencia, A. Implementing the iHOP concept for navigation of biomedical literature. Bioinformatics 21 (Suppl. 2), ii252?ii258 (2005).
Liebel, U., Kindler, B. & Pepperkok, R. 'Harvester': a fast meta search engine of human protein resources. Bioinformatics 20, 1962?1963 (2004).
Tong, A. H. et al. Global mapping of the yeast genetic interaction network. Science 303, 808?813 (2004).
DasGupta, R., Kaykas, A., Moon, R. T. & Perrimon, N. Functional genomic analysis of the Wnt?Wingless signaling pathway. Science 308, 826?833 (2005).
Huang, D., Moffat, J. & Andrews, B. Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol. Cell. Biol. 22, 5076?5088 (2002).
Sarbassov, D. D., Ali, S. M. & Sabatini, D. M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17, 596?603 (2005).
Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. Phosphorylation and regulation of Akt/PKB by the rictor?mTOR complex. Science 307, 1098?1101 (2005).
Matsuda, T. & Cepko, C. L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl Acad. Sci. USA 101, 16?22 (2004).
Hommel, J. D., Sears, R. M., Georgescu, D., Simmons, D. L. & DiLeone, R. J. Local gene knockdown in the brain using viral-mediated RNA interference. Nature Med. 9, 1539?1544 (2003).
Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. siRNA-mediated gene silencing in vitro and in vivo. Nature Biotechnol. 20, 1006?1010 (2002).
Kishida, T. et al. Sequence-specific gene silencing in murine muscle induced by electroporation-mediated transfer of short interfering RNA. J. Gene Med. 6, 105?110 (2004).
Kunath, T. et al. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nature Biotechnol. 21, 559?561 (2003).
Ventura, A. et al. Cre?lox-regulated conditional RNA interference from transgenes. Proc. Natl Acad. Sci. USA 101, 10380?10385 (2004).
Tiscornia, G., Singer, O., Ikawa, M. & Verma, I. M. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl Acad. Sci. USA 100, 1844?1848 (2003).
Rubinson, D. A. et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature Genet. 33, 401?406 (2003).
Siolas, D. et al. Synthetic shRNAs as potent RNAi triggers. Nature Biotechnol. 23, 227?231 (2005).
Carpenter, A. E. & Sabatini, D. M. Systematic genome-wide screens of gene function. Nature Rev. Genet. 5, 11?22 (2004).
Acknowledgements
We thank K. Ottina, J. Reiling, C. Thoreen and D. Guertin for helpful comments and critical reading of the manuscript and all laboratory members for valuable discussions. We apologize to authors whose primary works have not been cited due to space constraints. The work of the authors is supported by a Natural Sciences and Engineering Research Council of Canada postdoctoral fellowship (J.M.) and grants from the National Institutes of Health, Keck Foundation and Stewart Trust (D.M.S.). The authors would also like to acknowledge members of the RNAi Consortium (http://www.broad.mit.edu/rnai_platform/) for their continued support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Small interfering RNA
-
(siRNA). A class of 19?22-nucleotide-long RNA molecules that interfere with the expression of genes by eliciting the RNAi response. siRNAs are short double-stranded RNA molecules with 2-nucleotide overhangs on either end, including a 5′ phosphate group and a 3′ hydroxyl group. They can be artificially introduced into cells to bring about the knockdown of a particular gene.
- Interferon response
-
A primitive antiviral response to dsRNAs of >30 base pairs, which triggers the sequence-nonspecific degradation of mRNA and the downregulation of cellular protein synthesis.
- Short hairpin RNA
-
(shRNA). A short RNA that contains sense and antisense sequences from a target gene that are connected by a hairpin loop. shRNAs can be expressed from a pol-III-type promoter or in the context of a microRNA by pol II promoters. Following processing of the shRNAs, the resulting siRNAs can decrease the expression of a gene that has complementary sequences by RNAi.
- MicroRNA
-
(miRNA). A small non-coding RNA of 19?25 nucleotides in length that regulates the expression of genes at the stage of protein synthesis.
- Reverse transfection
-
A process whereby cells are transfected with features (for example, DNA or RNA) that are immobilized on glass slides or in multi-well plates.
- RNA polymerase III promoter
-
A promoter that uses RNA pol III to drive the production of 5S RNA, tRNA and other small RNAs. U6 and H1 pol III promoters have all the elements that are required for the initiation of transcription upstream of a defined start site and the termination of transcription at four or more Ts.
- Primary polyadenylated RNAs
-
(pri-miRNA). A long primary polyadenylated miRNA that is transcribed by RNA pol II. The miRNA sequence and its reverse complement base pair to form a dsRNA hairpin loop, which forms the primary RNA structure.
- Microprocessor complex
-
A small protein complex consisting of Drosha and DGCR8 that is necessary and sufficient for mediating the genesis of miRNAs from the primary miRNA transcript.
- Pre-miRNA
-
A miRNA precursor that is converted from the pri-miRNA in the nucleus by the Microprocessor complex and exported to the cytoplasm by a mechanism that is mediated by exportin-5. The Dicer enzyme then cuts 20?25 nucleotides from the base of the hairpin to release the mature miRNA.
- High-content image-based screen
-
(HCS). A method that uses high-resolution images as the readout for a screen. This type of screening is typically carried out using automated microscopy to acquire images. The images are analysed by eye or by automated image analysis, which is sometimes referred to as HCA (high-content analysis).
- Epistasis
-
The masking of a phenotype that is caused by a mutation in one gene, by a mutation in another gene. Epistasis analysis can therefore be used to dissect the order in which genes function in a genetic pathway.
- Perturbagen
-
A reagent (for example, a chemical or siRNA) or condition that disrupts or modifies the function of a specific gene or signalling pathway.
- Cell microarray
-
A method for studying cells that take up perturbagens and that have been printed in an arrayed format on the surface of glass slides.
- Reverse genetics
-
Genetic analysis that proceeds from genotype to phenotype through gene-manipulation techniques.
- Blastocyst
-
The stage of the developing embryo when the number of cells reaches 40?150, a central fluid-filled cavity called the blastocoel forms and the zona pellucida begins to degenerate. This stage lasts approximately until implantation into the uterus.
- Tetraploid aggregation
-
A method that is used to generate embryos that are completely derived from embryonic stem cells. The approach provides a quick evaluation of phenotype in embryonic development; for example, one can observe a knockout phenotype by aggregating mouse embryos with gene-knockout or transgenic RNAi embryonic stem cells.
- Perivitelline space
-
Region between the surface of the oocyte or more specifically the oolemma and the zona pellucida, an extracellular matrix synthesized by the oocyte. The perivitelline space has contents that change during development and that appear to have various roles before, during and after fertilization.
Rights and permissions
About this article
Cite this article
Moffat, J., Sabatini, D. Building mammalian signalling pathways with RNAi screens. Nat Rev Mol Cell Biol 7, 177–187 (2006). https://doi.org/10.1038/nrm1860
Issue Date:
DOI: https://doi.org/10.1038/nrm1860
This article is cited by
-
ER-mitochondria contact sites in neurodegeneration: genetic screening approaches to investigate novel disease mechanisms
Cell Death & Differentiation (2021)
-
Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening
Nature Protocols (2017)
-
Method for Dual Viral Vector Mediated CRISPR-Cas9 Gene Disruption in Primary Human Endothelial Cells
Scientific Reports (2017)
-
Identifying the topology of signaling networks from partial RNAi data
BMC Systems Biology (2016)
-
An integrative approach for a network based meta-analysis of viral RNAi screens
Algorithms for Molecular Biology (2015)