Abstract
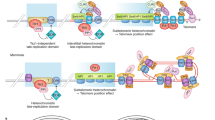
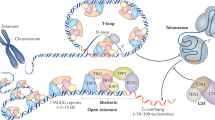
Most eukaryotes stabilize the ends of their linear chromosomes with a telomerase-based system. Telomerase maintains specific repetitive sequences, which protect chromosome ends with the help of telomere-binding proteins. How did this elaborate system evolve? Here, I propose that telomere function was originally mediated by t-loops, which could have been generated by prokaryotic DNA-replication factors. These early telomeres would have required only the presence of a few repeats at chromosome ends. Telomerase could have been a later innovation with specific advantages for telomere function and regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Greider, C. W. & Blackburn, E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413 (1985).
Nugent, C. I. & Lundblad, V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 12, 1073–1085 (1998).
Lundblad, V. Telomerase catalysis: a phylogenetically conserved reverse transcriptase. Proc. Natl Acad. Sci. USA 95, 8415–8416 (1998).
Kobryn, K. & Chaconas, G. The circle is broken: telomere resolution in linear replicons. Curr. Opin. Microbiol. 4, 558–564 (2001).
Varmus, H. E. Form and function of retroviral proviruses. Science 216, 812–820 (1982).
de Jong, R. N., van der Vliet, P. C. & Brenkman, A. B. Adenovirus DNA replication: protein priming, jumping back and the role of the DNA binding protein DBP. Curr. Top. Microbiol. Immunol. 272, 187–211 (2003).
Biessmann, H. & Mason, J. M. Telomere maintenance without telomerase. Chromosoma 106, 63–69 (1997).
Frydrychova, R. & Marec, F. Repeated losses of TTAGG telomere repeats in evolution of beetles (Coleoptera). Genetica 115, 179–187 (2002).
Lundblad, V. & Szostak, J. W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57, 633–643 (1989).
Lundblad, V. & Blackburn, E. H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73, 347–360 (1993).
Kass-Eisler, A. & Greider, C. W. Recombination in telomere-length maintenance. Trends Biochem. Sci. 25, 200–204 (2000).
Natarajan, S. & McEachern, M. J. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22, 4512–4521 (2002).
Bryan, T. M., Englezou, A., Dalla-Pozza, L., Dunham, M. A. & Reddel, R. R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nature Med. 3, 1271–1274 (1997).
Henson, J. D., Neumann, A. A., Yeager, T. R. & Reddel, R. R. Alternative lengthening of telomeres in mammalian cells. Oncogene 21, 598–610 (2002).
de Lange, T. Protection of mammalian telomeres. Oncogene 21, 532–540 (2002).
Lustig, A. J. Cdc13 subcomplexes regulate multiple telomere functions. Nature Struct. Biol. 8, 297–299 (2001).
Ferreira, M. G., Miller, K. M. & Cooper, J. P. Indecent exposure. When telomeres become uncapped. Mol. Cell 13, 7–18 (2004).
Singer, M. S. & Gottschling, D. E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266, 404–409 (1994).
Blasco, M. A. et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34 (1997).
Gottschling, D. E. & Zakian, V. A. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell 47, 195–205 (1986).
Griffith, J. D. et al. Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 (1999).
Stansel, R. M., de Lange, T. & Griffith, J. D. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20, E5532–5540 (2001).
de Lange, T. & Petrini, J. A new connection at human telomeres: association of the Mre11 complex with TRF2. Cold Spring Harb. Symp. Quant. Biol. LXV, 265–273 (2000).
Opresko, P. L. et al. Telomere binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 277, 41110–41119 (2002).
Sun, H., Karow, J. K., Hickson, I. D. & Maizels, N. P. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273, 27587–27592 (1998).
Sun, H., Bennett, R. J. & Maizels, N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G–G paired DNAs. Nucleic Acids Res. 27, 1978–1984 (1999).
Lustig, A. J. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nature Rev. Genet. 4, 916–923 (2003).
Mosig, G. The essential role of recombination in phage T4 growth. Annu. Rev. Genet. 21, 347–371 (1987).
Kreuzer, K. N. Recombination-dependent DNA replication in phage T4. Trends Biochem. Sci. 25, 165–173 (2000).
Cox, M. M. et al. The importance of repairing stalled replication forks. Nature 404, 37–41 (2000).
Wei, C., Skopp, R., Takata, M., Takeda, S. & Price, C. M. Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res. 30, 2862–2870 (2002).
Jaco, I. et al. Role of mammalian Rad54 in telomere length maintenance. Mol. Cell. Biol. 23, 5572–5580 (2003).
Le, S., Moore, J. K., Haber, J. E. & Greider, C. W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152, 143–152 (1999).
Goldbach, R. W., Bollen-de Boer, J. E., van Bruggen, E. F. & Borst, P. Replication of the linear mitochondrial DNA of Tetrahymena pyriformis. Biochim. Biophys. Acta 562, 400–417 (1979).
Morin, G. B. & Cech, T. R. The telomeres of the linear mitochondrial DNA of Tetrahymena thermophila consist of 53 bp tandem repeats. Cell 46, 873–883 (1986).
Morin, G. B. & Cech, T. R. Mitochondrial telomeres: surprising diversity of repeated telomeric DNA sequences among six species of Tetrahymena. Cell 52, 367–374 (1988).
Cech, T. R., Nakamura, T. M. & Lingner, J. Telomerase is a true reverse transcriptase. A review. Biochemistry (Mosc.) 62, 1202–1205 (1997).
Eickbush, T. H. Telomerase and retrotransposons: which came first? Science 277, 911–912 (1997).
Klobutcher, L. A., Swanton, M. T., Donini, P. & Prescott, D. M. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl Acad. Sci. USA 78, 3015–3019 (1981).
Lipps, H. J., Gruissem, W. & Prescott, D. M. Higher order DNA structure in macronuclear chromatin of the hypotrichous ciliate Oxytricha nova. Proc. Natl Acad. Sci. USA 79, 2495–2499 (1982).
Gottschling, D. E. & Cech, T. R. Chromatin structure of the molecular ends of Oxytricha macronuclear DNA: phased nucleosomes and a telomeric complex. Cell 38, 501–510 (1984).
Lingner, J. et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276, 561–567 (1997).
Smogorzewska, A. & de Lange, T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. (in the press).
Murti, K. G. & Prescott, D. M. Telomeres of polytene chromosomes in a ciliated protozoan terminate in duplex DNA loops. Proc. Natl Acad. Sci. USA 96, 14436–14439 (1999).
Munoz-Jordan, J. L., Cross, G. A., de Lange, T. & Griffith, J. D. T-loops at trypanosome telomeres. Embo J. 20, 579–588 (2001).
Cesare, A. J., Quinney, N., Willcox, S., Subramanian, D. & Griffith, J. D. Telomere looping in P. sativum (common garden pea). Plant J. 36, 271–279 (2003).
Horvath, M. P., Schweiker, V. L., Bevilacqua, J. M., Ruggles, J. A. & Schultz, S. C. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95, 963–974 (1998).
Acknowledgements
I dedicate this article to the memory of Anat Krauskopf. K. Kreuzer, J. Haber, V. Lundblad, R. Reddel, K. Hoke, and S. Buonomo provided extremely helpful criticism of these ideas. I thank J. Griffith for comments on this manuscript and for long-term collaborative efforts on t-loops. B. Bowerman is acknowledged for sage advice ('Evolution happened. Get over it.'). Space limitations prevented me from providing a thorough review of all relevant publications and I refer to the cited reviews for many important primary references. Research on telomeres in my laboratory is funded by the National Institutes of Health, the National Cancer Institute and the Ellison Foundation.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
de Lange, T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol 5, 323–329 (2004). https://doi.org/10.1038/nrm1359
Issue Date:
DOI: https://doi.org/10.1038/nrm1359
This article is cited by
-
Telomere sequence variability in genotypes from natural plant populations: unusual block-organized double-monomer terminal telomeric arrays
BMC Genomics (2023)
-
The role of the DFF40/CAD endonuclease in genomic stability
Apoptosis (2021)
-
Association study of leukocyte telomere length and genetic polymorphism within hTERT promoter with type 2 diabetes in Bangladeshi population
Molecular Biology Reports (2021)
-
The regulation and functions of DNA and RNA G-quadruplexes
Nature Reviews Molecular Cell Biology (2020)
-
Systematics for types and effects of DNA variations
BMC Genomics (2018)