Key Points
-
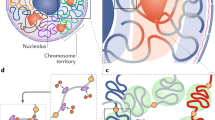
Contacts between distant genomic regions in the same or in different chromosomes are important in the regulation of gene expression, as highlighted by the activation of genes by chromatin contacts between promoters and enhancers that can lie hundreds of kb away.
-
Chromatin contacts are currently measured by two main approaches: chromosome conformation capture (3C)-based techniques and nuclear imaging methods such as fluorescence in situ hybridization (FISH). Both approaches have caveats, and the field is ripe for further technical development.
-
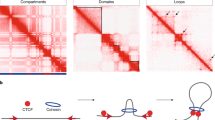
The formation of chromatin contacts is promoted by chromatin-binding proteins that can bind two or more genomic regions simultaneously. Such proteins include transcription factors, RNA and DNA polymerases, Polycomb repressive complexes and chromosomal scaffold proteins such as cohesin.
-
Topologically associating domains (TADs) are genomic regions enriched with contacts within them. TADs have specific sizes and positions in the genome and are found in a wide range of metazoans. The factors and mechanisms that promote TAD formation are a matter of considerable debate.
-
The three-dimensional organization of the genome also depends on the formation of chromatin contacts with nuclear domains and compartments such as the nuclear lamina and the nucleolus. Specific sets of chromatin contacts are formed within each chromosome, and between them and nuclear domains. The mechanisms that govern chromosome localization, volume and shape remain poorly understood.
-
Many cellular processes such as division, differentiation and senescence, present challenges to the maintenance of nuclear organization, gene expression programs and cell identity. At the same time, they can also offer opportunities for chromatin remodelling and the reinforcement of gene expression patterns.
Abstract
The different cell types of an organism share the same DNA, but during cell differentiation their genomes undergo diverse structural and organizational changes that affect gene expression and other cellular functions. These can range from large-scale folding of whole chromosomes or of smaller genomic regions, to the re-organization of local interactions between enhancers and promoters, mediated by the binding of transcription factors and chromatin looping. The higher-order organization of chromatin is also influenced by the specificity of the contacts that it makes with nuclear structures such as the lamina. Sophisticated methods for mapping chromatin contacts are generating genome-wide data that provide deep insights into the formation of chromatin interactions, and into their roles in the organization and function of the eukaryotic cell nucleus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
05 August 2015
In the original article, the following sentence was incorrect: “Histone marks associated with enhancers, such as histone 3 Lys4 monomethylation (H3K4me1), and with transcription repression, such as histone 3 Lys9 trimethylation (H3K9me3), were also enriched at TAD boundaries.2” The corrected sentence is as follows: “Histone marks associated with enhancers, such as histone 3 Lys4 monomethylation (H3K4me1), and with transcription repression, such as histone 3 Lys9 trimethylation (H3K9me3), were not found to be enriched at TAD boundaries.” This has been corrected in the online version of the article.
References
Wijgerde, M., Grosveld, F. & Fraser, P. Transcription complex stability and chromatin dynamics in vivo. Nature 377, 209–213 (1995).
Dillon, N., Trimborn, T., Strouboulis, J., Fraser, P. & Grosveld, F. The effect of distance on long-range chromatin interactions. Mol. Cell 1, 131–139 (1997).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nature Genet. 38, 1348–1354 (2006).
Naka, K. & Hirao, A. Maintenance of genomic integrity in hematopoietic stem cells. Int. J. Hematol. 93, 434–439 (2011).
Stadhouders, R. et al. Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nature Protoc. 8, 509–524 (2013).
Dostie, J. et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Hughes, J. R. et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature Genet. 46, 205–212 (2014).
Rodley, C. D., Bertels, F., Jones, B. & O'Sullivan, J. M. Global identification of yeast chromosome interactions using genome conformation capture. Fungal Genet. Biol. 46, 879–886 (2009).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Kalhor, R., Tjong, H., Jayathilaka, N., Alber, F. & Chen, L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nature Biotech. 30, 90–98 (2012).
Kolovos, P. et al. Targeted Chromatin Capture (T2C): a novel high resolution high throughput method to detect genomic interactions and regulatory elements. Epigenet. Chromatin 7, 10 (2014).
Tolhuis, B., Palstra, R. J., Splinter, E., Grosveld, F. & de Laat, W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 (2002).
Vernimmen, D., De Gobbi, M., Sloane-Stanley, J. A., Wood, W. G. & Higgs, D. R. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 26, 2041–2051 (2007).
Stadhouders, R. et al. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 31, 986–999 (2012).
Jing, H. et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol. Cell 29, 232–242 (2008).
Markova, E. N., Kantidze, O. L. & Razin, S. V. Transcriptional regulation and spatial organisation of the human AML1/RUNX1 gene. J. Cell Biochem. 112, 1997–2005 (2011).
Blackledge, N. P., Ott, C. J., Gillen, A. E. & Harris, A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 37, 1086–1094 (2009).
Ktistaki, E. et al. CD8 locus nuclear dynamics during thymocyte development. J. Immunol. 184, 5686–5695 (2010).
Palstra, R. J. et al. The β-globin nuclear compartment in development and erythroid differentiation. Nature Genet. 35, 190–194 (2003).
Love, P. E., Warzecha, C. & Li, L. Ldb1 complexes: the new master regulators of erythroid gene transcription. Trends Genet. 30, 1–9 (2014).
Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014).
Grosveld, F., van Assendelft, G. B., Greaves, D. R. & Kollias, G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 51, 975–985 (1987).
Sabbattini, P., Georgiou, A., Sinclair, C. & Dillon, N. Analysis of mice with single copies and multiple copies of transgenes reveals a novel arrangement for the λ5-VpreB1 locus control region. Mol. Cell. Biol. 19, 671–679 (1999).
Fields, P. E., Lee, G. R., Kim, S. T., Bartsevich, V. V. & Flavell, R. A. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity 21, 865–876 (2004).
Ellis, J., Talbot, D., Dillon, N. & Grosveld, F. Synthetic human β-globin 5′HS2 constructs function as locus control regions only in multicopy transgene concatamers. EMBO J. 12, 127–134 (1993).
Nasmyth, K. & Haering, C. H. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 (2009).
Parelho, V. et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 (2008).
Hadjur, S. et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 (2009).
Seitan, V. C. et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476, 467–471 (2011).
Lobanenkov, V. V. et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5, 1743–1753 (1990).
Ong, C. T. & Corces, V. G. CTCF: an architectural protein bridging genome topology and function. Nature Rev. Genet. 15, 234–246 (2014).
Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Xu, Z., Wei, G., Chepelev, I., Zhao, K. & Felsenfeld, G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nature Struct. Mol. Biol. 18, 372–378 (2011).
Kehayova, P., Monahan, K., Chen, W. & Maniatis, T. Regulatory elements required for the activation and repression of the protocadherin-α gene cluster. Proc. Natl Acad. Sci. USA 108, 17195–17200 (2011).
Guo, Y. et al. CTCF/cohesin-mediated DNA looping is required for protocadherin-α promoter choice. Proc. Natl Acad. Sci. USA 109, 21081–21086 (2012).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Hou, C., Li, L., Qin, Z. S. & Corces, V. G. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48, 471–484 (2012).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Van Bortle, K. et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 15, R82 (2014).
Kim, Y. J., Cecchini, K. R. & Kim, T. H. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc. Natl Acad. Sci. USA 108, 7391–7396 (2011).
Seitan, V. C. et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 23, 2066–2077 (2013).
Zuin, J. et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl Acad. Sci. USA 111, 996–1001 (2014).
Sofueva, S. et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 32, 3119–3129 (2013).
Young, S. G., Jung, H. J., Coffinier, C. & Fong, L. G. Understanding the roles of nuclear A- and B-type lamins in brain development. J. Biol. Chem. 287, 16103–16110 (2012).
Houben, F. et al. Disturbed nuclear orientation and cellular migration in A-type lamin deficient cells. Biochim. Biophys. Acta 1793, 312–324 (2009).
Dechat, T., Adam, S. A., Taimen, P., Shimi, T. & Goldman, R. D. Nuclear lamins. CSH Persp. Biol. 2, a000547 (2010).
Amendola, M. & van Steensel, B. Mechanisms and dynamics of nuclear lamina-genome interactions. Curr. Opin. Cell Biol. 28, 61–68 (2014).
Guelen, L. et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008).
Meuleman, W. et al. Constitutive nuclear lamina–genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 23, 270–280 (2013).
Finlan, L. E. et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4, e1000039 (2008).
Lundgren, M. et al. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103, 733–743 (2000).
Kind, J. et al. Single-cell dynamics of genome–nuclear lamina interactions. Cell 153, 178–192 (2013).
Padeken, J. & Heun, P. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr. Opin. Cell Biol. 28, 54–60 (2014).
van Koningsbruggen, S. et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell 21, 3735–3748 (2010).
Nemeth, A. et al. Initial genomics of the human nucleolus. PLoS Genet. 6, e1000889 (2010).
Parada, L. A., McQueen, P. G. & Misteli, T. Tissue-specific spatial organization of genomes. Genome Biol. 5, R44 (2004).
Tanabe, H. et al. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl Acad. Sci. USA 99, 4424–4429 (2002).
Bridger, J. M. Chromobility: the rapid movement of chromosomes in interphase nuclei. Biochem. Soc. Trans. 39, 1747–1751 (2011).
Williams, R. R., Broad, S., Sheer, D. & Ragoussis, J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272, 163–175 (2002).
Ferrai, C. et al. Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol. 8, e1000270 (2010).
Volpi, E. et al. Large-scale chromatin organisation of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei J. Cell Sci. 113, 1565–1576 (2000).
Meaburn, K. J., Gudla, P. R., Khan, S., Lockett, S. J. & Misteli, T. Disease-specific gene repositioning in breast cancer. J. Cell Biol. 187, 801–812 (2009).
Cremer, T. et al. Chromosome territories — a functional nuclear landscape. Curr. Opin. Cell Biol. 18, 307–316 (2006).
Branco, M. R. & Pombo, A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4, e138 (2006).
Branco, M. R., Branco, T., Ramirez, F. & Pombo, A. Changes in chromosome organization during PHA-activation of resting human lymphocytes measured by cryo-FISH. Chromosome Res. 16, 413–426 (2008).
Zhang, Y. et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148, 908–921 (2012).
Hakim, O. et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 484, 69–74 (2012).
Roukos, V. et al. Spatial dynamics of chromosome translocations in living cells. Science 341, 660–664 (2013).
Jackson, D. A. & Pombo, A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295 (1998).
Ma, H. et al. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 143, 1415–1425 (1998).
Gottesfeld, J. M. & Forbes, D. J. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197–202 (1997).
Belmont, A. S. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 18, 632–638 (2006).
Naumova, N. et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013).
Chow, C.-M. et al. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 6, 354–360 (2005).
Kouskouti, A. & Talianidis, I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24, 347–357 (2004).
Kelly, T. K. et al. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol. Cell 39, 901–911 (2010).
Caravaca, J. M. et al. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 27, 251–260 (2013).
Kadauke, S. et al. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150, 725–737 (2012).
Young, D. W. et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc. Natl Acad. Sci. USA 104, 3189–3194 (2007).
Blobel, G. A. et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell 36, 970–983 (2009).
Zhao, R., Nakamura, T., Fu, Y., Lazar, Z. & Spector, D. L. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nature Cell Biol. 13, 1295–1304 (2011).
Rawlings, J. S., Gatzka, M., Thomas, P. G. & Ihle, J. N. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 30, 263–276 (2011).
Sabbattini, P. et al. An H3K9/S10 methyl-phospho switch modulates Polycomb and Pol II binding at repressed genes during differentiation. Mol. Biol. Cell 25, 904–915 (2014).
Frangini, A. et al. The Aurora B kinase and the polycomb protein Ring1B combine to regulate active promoters in quiescent lymphocytes. Mol. Cell 51, 647–661 (2013).
Sabbattini, P. et al. A novel role for the Aurora B kinase in epigenetic marking of silent chromatin in differentiated postmitotic cells. EMBO J. 26, 4657–4669 (2007).
Fischle, W. et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005).
Solovei, I. et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 (2013).
Solovei, I. et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137, 356–368 (2009).
Helmlinger, D. et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 4, e67 (2006).
Rai, T. S. & Adams, P. D. Lessons from senescence: chromatin maintenance in non-proliferating cells. Biochim. Biophys. Acta 1819, 322–331 (2013).
Zhang, R., Chen, W. & Adams, P. D. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2343–2358 (2007).
Ye, X. et al. Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol. Cell 27, 183–196 (2007).
Freund, A., Laberge, R. M., Demaria, M. & Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 23, 2066–2075 (2012).
Shimi, T. et al. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 25, 2579–2593 (2011).
Shah, P. P. et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 27, 1787–1799 (2013).
Sadaie, M. et al. Redistribution of the lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 27, 1800–1808 (2013).
Friedl, P., Wolf, K. & Lammerding, J. Nuclear mechanics during cell migration. Curr. Opin. Cell Biol. 23, 55–64 (2011).
Mohrin, M. et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185 (2010).
Rehen, S. K. et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl Acad. Sci. USA 98, 13361–13366 (2001).
Duncan, A. W. et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 467, 707–710 (2010).
Terns, R. M. & Terns, M. P. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet. 30, 111–118 (2014).
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nature Rev. Genet. 14, 390–403 (2013).
Ethier, S. D., Miura, H. & Dostie, J. Discovering genome regulation with 3C and 3C-related technologies. Biochim. Biophys. Acta 1819, 401–410 (2012).
de Wit, E. & de Laat, W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 (2012).
van de Werken, H. J. et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nature Methods 9, 969–972 (2012).
Belmont, A. S. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr. Opin. Cell Biol. 26, 69–78 (2014).
Gavrilov, A. A. et al. Disclosure of a structural milieu for the proximity ligation reveals the elusive nature of an active chromatin hub. Nucleic Acids Res. 41, 3563–3575 (2013).
O'Sullivan, J. M. Hendy, M. D., Pichugina, T., Wake, G. C. & Langowski, J. The statistical-mechanics of chromosome conformation capture. Nucleus 4, 390–398 (2013).
Nicodemi, M. & Pombo, A. Models of chromosome structure. Curr. Opin. Cell Biol. 28, 90–95 (2014).
Barbieri, M. et al. Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl Acad. Sci. USA 109, 16173–16178 (2012).
Bau, D. et al. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nature Struct. Mol. Biol. 18, 107–114 (2011).
Nagano, T. et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013).
Hozak, P. & Cook, P. R. Replication factories. Trends Cell Biol. 4, 48–52 (1994).
Baddeley, D. et al. Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res. 38, e8 (2010).
Jackson, D. A., Iborra, F. J., Manders, E. M. & Cook, P. R. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 9, 1523–1536 (1998).
Martin, S. & Pombo, A. Transcription factories: quantitative studies of nanostructures in the mammalian nucleus. Chromosome Res. 11, 461–470 (2003).
Pombo, A. et al. Specialized transcription factories within mammalian nuclei. Crit. Rev. Eukaryot. Gene Expr 10, 21–29 (2000).
Kimura, H., Tao, Y., Roeder, R. G. & Cook, P. R. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell. Biol. 19, 5383–5392 (1999).
Pombo, A. et al. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 18, 2241–2253 (1999).
Faro-Trindade, I. & Cook, P. R. A conserved organization of transcription during embryonic stem cell differentiation and in cells with high C value. Mol. Biol. Cell 17, 2910–2920 (2006).
Jackson, D. A. Features of nuclear architecture that influence gene expression in higher eukaryotes: confronting the enigma of epigenetics. J. Cell Biochem. 79(Suppl.35), 69–77 (2000).
Osborne, C. et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature Genet. 36, 1065–1071 (2004).
Schoenfelder, S. et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature Genet. 42, 53–61 (2010).
Brookes, E. et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10, 157–170 (2012).
Lanzuolo, C., Roure, V., Dekker, J., Bantignies, F. & Orlando, V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nature Cell Biol. 9, 1167–1174 (2007).
Grimaud, C. et al. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell 124, 957–971 (2006).
Tiwari, V. K., Cope, L., McGarvey, K. M., Ohm, J. E. & Baylin, S. B. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 18, 1171–1179 (2008).
Acknowledgements
The authors thank P. Sabbattini for providing the images shown in Fig. 4a. Work in N.D.'s laboratory is supported by the Medical Research Council, UK. A.P. thanks the Helmholtz Foundation for support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (box)
Relating chromatin contacts to gene regulation mechanisms (PDF 117 kb)
Glossary
- Chromatin immunoprecipitation
-
A method in which chromatin bound by a protein is immunoprecipitated with an antibody against that protein, to allow the extraction and analysis of the bound DNA by quantitative PCR or genome-wide sequencing.
- Nuclear lamina
-
A protein meshwork made of intermediate filaments (such as lamins) and membrane-associated proteins (such as emerin) that covers the inner nuclear membrane and is responsible for maintaining nuclear shape, organization and function.
- Heterochromatin
-
Highly condensed chromatin that shows dark staining. Constitutive heterochromatin remains in this state throughout the cell cycle. Facultative heterochromatin is cell-type-specific condensed chromatin that is often a feature of terminally differentiated cells.
- DNA adenine methyltransferase identification
-
A method based on expression of fusion proteins with bacterial Dam methylase, and detection of methylated DNA as a measure of its contact with the fusion protein.
- Cryoprotected cells
-
Cells that have been treated with a cryoproctectant to prevent structural damage during freezing.
Rights and permissions
About this article
Cite this article
Pombo, A., Dillon, N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16, 245–257 (2015). https://doi.org/10.1038/nrm3965
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3965
This article is cited by
-
Site-specific transgene integration in chimeric antigen receptor (CAR) T cell therapies
Biomarker Research (2023)
-
Auxin-inducible degron 2 system deciphers functions of CTCF domains in transcriptional regulation
Genome Biology (2023)
-
Enhancer–promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness
Nature Genetics (2023)
-
TAD evolutionary and functional characterization reveals diversity in mammalian TAD boundary properties and function
Nature Communications (2023)
-
Contribution of 3D genome topological domains to genetic risk of cancers: a genome-wide computational study
Human Genomics (2022)