Key Points
-
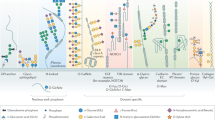
Protein glycosylation describes various conserved post-translational modifications. Although some proteins can be glycosylated in the cytosol, glycosylation is most prevalent in the secretory pathway, in which it is involved in many processes.
-
Proteins that are synthesized at the endoplasmic reticulum (ER) membranes can be glycosylated at Asn, Ser and Thr residues. These can be further modified in the Golgi apparatus.
-
The core N-linked glycan has a branched structure composed of Glc3Man9GlcNAc2, which is trimmed sequentially by ER glycosidases. The trimming events coordinate folding and quality control factors to ensure high-fidelity production of mature glycoproteins.
-
Trimming of an N-linked glycan to the Man7GlcNAc2 structure bearing a terminal α-1,6-linked mannose residue attached to an unfolded peptide allows peptide recognition by the yeast osteosarcoma 9 (Yos9) ER-associated degradation (ERAD) receptor. This initiates peptide retro-translocation and degradation by the ubiquitin–proteasome system.
-
In budding yeast, misfolded proteins are modified by O-mannosylation by the Pmt1–Pmt2 complex. This modification can facilitate ERAD or promote the transport of the substrate out of the ER to the vacuole or cell surface.
-
O-mannosylation is used in budding yeast to terminate futile cycles of protein folding to facilitate their entry into ERAD and relieve ER stress.
Abstract
Membrane-bound and soluble proteins of the secretory pathway are commonly glycosylated in the endoplasmic reticulum. These adducts have many biological functions, including, notably, their contribution to the maturation of glycoproteins. N-linked glycans are of oligomeric structure, forming configurations that provide blueprints to precisely instruct the folding of protein substrates and the quality control systems that scrutinize it. O-linked mannoses are simpler in structure and were recently found to have distinct functions in protein quality control that do not require the complex structure of N-linked glycans. Together, recent studies reveal the breadth and sophistication of the roles of these glycan-directed modifications in protein biogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Park, E. & Rapoport, T. A. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 (2012).
Gidalevitz, T., Stevens, F. & Argon, Y. Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta 1833, 2410–2424 (2013).
Braakman, I. & Hebert, D. N. Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, a013201 (2013).
Araki, K. & Nagata, K. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 4, a015438 (2012).
Kelleher, D. J. & Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R (2006).
Lizak, C., Gerber, S., Numao, S., Aebi, M. & Locher, K. P. X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 (2011).
Ruiz-Canada, C., Kelleher, D. J. & Gilmore, R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136, 272–283 (2009).
Helenius, J. et al. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415, 447–450 (2002).
Breitling, J. & Aebi, M. N-linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, a013359 (2013).
Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437 (2013).
Takeuchi, H. & Haltiwanger, R. S. Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 453, 235–242 (2014).
Sentandreu, R. & Northcote, D. H. The structure of a glycopeptide isolated from the yeast cell wall. Biochem. J. 109, 419–432 (1968).
Marriott, M. & Tanner, W. Localization of dolichyl phosphate- and pyrophosphate-dependent glycosyl transfer reactions in Saccharomyces cerevisiae. J. Bacteriol. 139, 566–572 (1979).
Haselbeck, A. & Tanner, W. O-glycosylation in Saccharomyces cerevisiae is initiated at the endoplasmic reticulum. FEBS Lett. 158, 335–338 (1983).
Maeda, Y. & Kinoshita, T. Dolichol-phosphate mannose synthase: structure, function and regulation. Biochim. Biophys. Acta 1780, 861–868 (2008).
Strahl-Bolsinger, S., Immervoll, T., Deutzmann, R. & Tanner, W. PMT1, the gene for a key enzyme of protein O-glycosylation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 90, 8164–8168 (1993).
Strahl-Bolsinger, S. & Tanner, W. Protein O-glycosylation in Saccharomyces cerevisiae. Purification and characterization of the dolichyl-phosphate-d-mannose-protein O-d-mannosyltransferase. Eur. J. Biochem. 196, 185–190 (1991).
Praissman, J. L. & Wells, L. Mammalian O-mannosylation pathway: glycan structures, enzymes, and protein substrates. Biochemistry 53, 3066–3078 (2014).
Loibl, M. & Strahl, S. Protein O-mannosylation: what we have learned from baker's yeast. Biochim. Biophys. Acta 1833, 2438–2446 (2013).
Girrbach, V. & Strahl, S. Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 278, 12554–12562 (2003).
Girrbach, V., Zeller, T., Priesmeier, M. & Strahl-Bolsinger, S. Structure-function analysis of the dolichyl phosphate-mannose: protein O-mannosyltransferase ScPmt1p. J. Biol. Chem. 275, 19288–19296 (2000).
Willer, T., Amselgruber, W., Deutzmann, R. & Strahl, S. Characterization of POMT2, a novel member of the PMT protein O-mannosyltransferase family specifically localized to the acrosome of mammalian spermatids. Glycobiology 12, 771–783 (2002).
Gentzsch, M., Immervoll, T. & Tanner, W. Protein O-glycosylation in Saccharomyces cerevisiae: the protein O-mannosyltransferases Pmt1p and Pmt2p function as heterodimer. FEBS Lett. 377, 128–130 (1995).
Akasaka-Manya, K., Manya, H., Nakajima, A., Kawakita, M. & Endo, T. Physical and functional association of human protein O-mannosyltransferases 1 and 2. J. Biol. Chem. 281, 19339–19345 (2006).
Rubenstein, E. M., Kreft, S. G., Greenblatt, W., Swanson, R. & Hochstrasser, M. Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. J. Cell Biol. 197, 761–773 (2012).
Harty, C., Strahl, S. & Romisch, K. O-mannosylation protects mutant alpha-factor precursor from endoplasmic reticulum-associated degradation. Mol. Biol. Cell 12, 1093–1101 (2001).
Vashist, S. et al. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 155, 355–368 (2001).
Nakatsukasa, K. et al. Roles of O-mannosylation of aberrant proteins in reduction of the load for endoplasmic reticulum chaperones in yeast. J. Biol. Chem. 279, 49762–49772 (2004).
Li, S., Spooner, R. A., Hampton, R. Y., Lord, J. M. & Roberts, L. M. Cytosolic entry of Shiga-like toxin A chain from the yeast endoplasmic reticulum requires catalytically active Hrd1p. PLoS ONE 7, e41119 (2012).
Hirayama, H., Fujita, M., Yoko-o, T. & Jigami, Y. O-mannosylation is required for degradation of the endoplasmic reticulum-associated degradation substrate Gas1*p via the ubiquitin/proteasome pathway in Saccharomyces cerevisiae. J. Biochem. 143, 555–567 (2008).
Coughlan, C. M., Walker, J. L., Cochran, J. C., Wittrup, K. D. & Brodsky, J. L. Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J. Biol. Chem. 279, 15289–15297 (2004).
Duttler, S., Pechmann, S. & Frydman, J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell 50, 379–393 (2013).
Ruggiano, A., Foresti, O. & Carvalho, P. Quality control: ER-associated degradation: protein quality control and beyond. J. Cell Biol. 204, 869–879 (2014).
Nakatsukasa, K., Kamura, T. & Brodsky, J. L. Recent technical developments in the study of ER-associated degradation. Curr. Opin. Cell Biol. 29, 82–91 (2014).
Christianson, J. C. & Ye, Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat. Struct. Mol. Biol. 21, 325–335 (2014).
Olzmann, J. A., Kopito, R. R. & Christianson, J. C. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5, a013185 (2013).
Thibault, G. & Ng, D. T. The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb. Perspect. Biol. 4, a013193 (2012).
Finger, A., Knop, M. & Wolf, D. H. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur. J. Biochem. 218, 565–574 (1993).
McCracken, A. A. & Brodsky, J. L. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132, 291–298 (1996).
Sommer, T. & Jentsch, S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature 365, 176–179 (1993).
Carvalho, P., Goder, V. & Rapoport, T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126, 361–373 (2006).
Vashist, S. & Ng, D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165, 41–52 (2004).
Nakatsukasa, K., Huyer, G., Michaelis, S. & Brodsky, J. L. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132, 101–112 (2008).
Habeck, G., Ebner, F. A., Shimada-Kreft, H. & Kreft, S. G. The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J. Cell Biol. 209, 261–273 (2015).
Machamer, C. E. & Rose, J. K. Vesicular stomatitis virus G proteins with altered glycosylation sites display temperature-sensitive intracellular transport and are subject to aberrant intermolecular disulfide bonding. J. Biol. Chem. 263, 5955–5960 (1988).
Gallagher, P., Henneberry, J., Wilson, I., Sambrook, J. & Gething, M. J. Addition of carbohydrate side chains at novel sites on influenza virus hemagglutinin can modulate the folding, transport, and activity of the molecule. J. Cell Biol. 107, 2059–2073 (1988).
Rutkevich, L. A. & Williams, D. B. Participation of lectin chaperones and thiol oxidoreductases in protein folding within the endoplasmic reticulum. Curr. Opin. Cell Biol. 23, 157–166 (2011).
Schrag, J. D., Procopio, D. O., Cygler, M., Thomas, D. Y. & Bergeron, J. J. Lectin control of protein folding and sorting in the secretory pathway. Trends Biochem. Sci. 28, 49–57 (2003).
Caramelo, J. J. & Parodi, A. J. Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 283, 10221–10225 (2008).
Ou, W. J., Cameron, P. H., Thomas, D. Y. & Bergeron, J. J. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature 364, 771–776 (1993).
Jackson, M. R., Cohen-Doyle, M. F., Peterson, P. A. & Williams, D. B. Regulation of MHC class I transport by the molecular chaperone, calnexin (p88, IP90). Science 263, 384–387 (1994).
Hammond, C., Braakman, I. & Helenius, A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl Acad. Sci. USA 91, 913–917 (1994).
Schrag, J. D. et al. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell 8, 633–644 (2001).
Molinari, M. & Helenius, A. Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature 402, 90–93 (1999).
Solda, T., Galli, C., Kaufman, R. J. & Molinari, M. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol. Cell 27, 238–249 (2007).
Tessier, D. C. et al. Cloning and characterization of mammalian UDP-glucose glycoprotein: glucosyltransferase and the development of a specific substrate for this enzyme. Glycobiology 10, 403–412 (2000).
Trombetta, E. S. & Helenius, A. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J. Cell Biol. 148, 1123–1129 (2000).
Ganan, S., Cazzulo, J. J. & Parodi, A. J. A major proportion of N-glycoproteins are transiently glucosylated in the endoplasmic reticulum. Biochemistry 30, 3098–3104 (1991).
Hitt, R. & Wolf, D. H. DER7, encoding α-glucosidase I is essential for degradation of malfolded glycoproteins of the endoplasmic reticulum. FEMS Yeast Res. 4, 815–820 (2004).
Camirand, A., Heysen, A., Grondin, B. & Herscovics, A. Glycoprotein biosynthesis in Saccharomyces cerevisiae. Isolation and characterization of the gene encoding a specific processing α-mannosidase. J. Biol. Chem. 266, 15120–15127 (1991).
Jakob, C. A., Burda, P., Roth, J. & Aebi, M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 142, 1223–1233 (1998).
Gauss, R., Kanehara, K., Carvalho, P., Ng, D. T. & Aebi, M. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol. Cell 42, 782–793 (2011).
Dancourt, J. & Barlowe, C. Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 79, 777–802 (2010).
Jakob, C. A. et al. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2, 423–430 (2001).
Movsichoff, F., Castro, O. A. & Parodi, A. J. Characterization of Schizosaccharomyces pombe ER α-mannosidase: a reevaluation of the role of the enzyme on ER-associated degradation. Mol. Biol. Cell 16, 4714–4724 (2005).
Munro, S. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Curr. Biol. 11, R499–R501 (2001).
Buschhorn, B. A., Kostova, Z., Medicherla, B. & Wolf, D. H. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett. 577, 422–426 (2004).
Bhamidipati, A., Denic, V., Quan, E. M. & Weissman, J. S. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell 19, 741–751 (2005).
Kim, W., Spear, E. D. & Ng, D. T. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol. Cell 19, 753–764 (2005).
Szathmary, R., Bielmann, R., Nita-Lazar, M., Burda, P. & Jakob, C. A. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell 19, 765–775 (2005).
Su, Y. A., Hutter, C. M., Trent, J. M. & Meltzer, P. S. Complete sequence analysis of a gene (OS-9) ubiquitously expressed in human tissues and amplified in sarcomas. Mol. Carcinog. 15, 270–275 (1996).
Quan, E. M. et al. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell 32, 870–877 (2008).
Clerc, S. et al. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184, 159–172 (2009).
Knop, M., Hauser, N. & Wolf, D. H. N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast 12, 1229–1238 (1996).
Nakatsukasa, K., Nishikawa, S., Hosokawa, N., Nagata, K. & Endo, T. Mnl1p, an α-mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J. Biol. Chem. 276, 8635–8638 (2001).
Xie, W., Kanehara, K., Sayeed, A. & Ng, D. T. Intrinsic conformational determinants signal protein misfolding to the Hrd1/Htm1 endoplasmic reticulum-associated degradation system. Mol. Biol. Cell 20, 3317–3329 (2009).
Sakoh-Nakatogawa, M., Nishikawa, S. & Endo, T. Roles of protein-disulfide isomerase-mediated disulfide bond formation of yeast Mnl1p in endoplasmic reticulum-associated degradation. J. Biol. Chem. 284, 11815–11825 (2009).
Nishikawa, S. I., Fewell, S. W., Kato, Y., Brodsky, J. L. & Endo, T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061–1070 (2001).
Buck, T. M., Kolb, A. R., Boyd, C. R., Kleyman, T. R. & Brodsky, J. L. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol. Biol. Cell 21, 1047–1058 (2010).
Kabani, M. et al. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell 14, 3437–3448 (2003).
Graham, T. R. & Emr, S. D. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J. Cell Biol. 114, 207–218 (1991).
Jungmann, J. & Munro, S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 17, 423–434 (1998).
Hirayama, H., Seino, J., Kitajima, T., Jigami, Y. & Suzuki, T. Free oligosaccharides to monitor glycoprotein endoplasmic reticulum-associated degradation in Saccharomyces cerevisiae. J. Biol. Chem. 285, 12390–12404 (2010).
Alonzi, D. S. et al. Glycoprotein misfolding in the endoplasmic reticulum: identification of released oligosaccharides reveals a second ER-associated degradation pathway for Golgi-retrieved proteins. Cell. Mol. Life Sci. 70, 2799–2814 (2013).
Mueller, B., Klemm, E. J., Spooner, E., Claessen, J. H. & Ploegh, H. L. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl Acad. Sci. USA 105, 12325–12330 (2008).
Christianson, J. C., Shaler, T. A., Tyler, R. E. & Kopito, R. R. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1–SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10, 272–282 (2008).
Gauss, R., Jarosch, E., Sommer, T. & Hirsch, C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat. Cell Biol. 8, 849–854 (2006).
Kanehara, K., Xie, W. & Ng, D. T. Modularity of the Hrd1 ERAD complex underlies its diverse client range. J. Cell Biol. 188, 707–716 (2010).
Mehnert, M. et al. The interplay of Hrd3 and the molecular chaperone system ensures efficient degradation of malfolded secretory proteins. Mol. Biol. Cell 26, 185–194 (2015).
Smith, M. H., Rodriguez, E. H. & Weissman, J. S. Misfolded proteins induce aggregation of the lectin Yos9. J. Biol. Chem. 289, 25670–25677 (2014).
Benitez, E. M., Stolz, A. & Wolf, D. H. Yos9, a control protein for misfolded glycosylated and non-glycosylated proteins in ERAD. FEBS Lett. 585, 3015–3019 (2011).
Horn, S. C. et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol. Cell 36, 782–793 (2009).
Taxis, C. et al. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 278, 35903–35913 (2003).
Mehnert, M., Sommer, T. & Jarosch, E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat. Cell Biol. 16, 77–86 (2014).
Lilley, B. N. & Ploegh, H. L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429, 834–840 (2004).
Scott, D. C. & Schekman, R. Role of Sec61p in the ER-associated degradation of short-lived transmembrane proteins. J. Cell Biol. 181, 1095–1105 (2008).
Pilon, M., Schekman, R. & Romisch, K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16, 4540–4548 (1997).
Walter, J., Urban, J., Volkwein, C. & Sommer, T. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 20, 3124–3131 (2001).
Schafer, A. & Wolf, D. H. Sec61p is part of the endoplasmic reticulum-associated degradation machinery. EMBO J. 28, 2874–2884 (2009).
Tretter, T. et al. ERAD and protein import defects in a sec61 mutant lacking ER-lumenal loop 7. BMC Cell Biol. 14, 56 (2013).
Carvalho, P., Stanley, A. M. & Rapoport, T. A. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 143, 579–591 (2010).
Biederer, T., Volkwein, C. & Sommer, T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278, 1806–1809 (1997).
Bays, N. W., Wilhovsky, S. K., Goradia, A., Hodgkiss-Harlow, K. & Hampton, R. Y. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12, 4114–4128 (2001).
Ye, Y., Meyer, H. H. & Rapoport, T. A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 (2001).
Jarosch, E. et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134–139 (2002).
Rabinovich, E., Kerem, A., Fröhlich, K. U., Diamant, N. & Bar-Nun, S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22, 626–634 (2002).
Stein, A., Ruggiano, A., Carvalho, P. & Rapoport, T. A. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell 158, 1375–1388 (2014).
Suzuki, T., Park, H., Hollingsworth, N. M., Sternglanz, R. & Lennarz, W. J. PNG1, a yeast gene encoding a highly conserved peptide: N-glycanase. J. Cell Biol. 149, 1039–1052 (2000).
Hirsch, C., Blom, D. & Ploegh, H. L. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22, 1036–1046 (2003).
Hirsch, C., Misaghi, S., Blom, D., Pacold, M. E. & Ploegh, H. L. Yeast N-glycanase distinguishes between native and non-native glycoproteins. EMBO Rep. 5, 201–206 (2004).
Garza, R. M., Sato, B. K. & Hampton, R. Y. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J. Biol. Chem. 284, 14710–14722 (2009).
Medicherla, B., Kostova, Z., Schaefer, A. & Wolf, D. H. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 5, 692–697 (2004).
Funakoshi, M., Sasaki, T., Nishimoto, T. & Kobayashi, H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl Acad. Sci. USA 99, 745–750 (2002).
Elsasser, S. et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4, 725–730 (2002).
Li, Y. et al. Rad4 regulates protein turnover at a postubiquitylation step. Mol. Biol. Cell 21, 177–185 (2009).
Ahner, A., Nakatsukasa, K., Zhang, H., Frizzell, R. A. & Brodsky, J. L. Small heat-shock proteins select ΔF508-CFTR for endoplasmic reticulum-associated degradation. Mol. Biol. Cell 18, 806–814 (2007).
Gonzalez, D. S., Karaveg, K., Vandersall-Nairn, A. S., Lal, A. & Moremen, K. W. Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J. Biol. Chem. 274, 21375–21386 (1999).
Avezov, E., Frenkel, Z., Ehrlich, M., Herscovics, A. & Lederkremer, G. Z. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5–6 GlcNAc2 in glycoprotein ER-associated degradation. Mol. Biol. Cell 19, 216–225 (2008).
Roth, J., Brada, D., Lackie, P. M., Schweden, J. & Bause, E. Oligosaccharide trimming Man9-mannosidase is a resident ER protein and exhibits a more restricted and local distribution than glucosidase II. Eur. J. Cell Biol. 53, 131–141 (1990).
Pan, S., Cheng, X. & Sifers, R. N. Golgi-situated endoplasmic reticulum α-1,2-mannosidase contributes to the retrieval of ERAD substrates through a direct interaction with γ-COP. Mol. Biol. Cell 24, 1111–1121 (2013).
Pan, S. et al. Golgi localization of ERManI defines spatial separation of the mammalian glycoprotein quality control system. Mol. Biol. Cell 22, 2810–2822 (2011).
Benyair, R. et al. Mammalian ER mannosidase I resides in quality control vesicles, where it encounters its glycoprotein substrates. Mol. Biol. Cell 26, 172–184 (2015).
Hosokawa, N. et al. A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2, 415–422 (2001).
Tamura, T., Cormier, J. H. & Hebert, D. N. Characterization of early EDEM1 protein maturation events and their functional implications. J. Biol. Chem. 286, 24906–24915 (2011).
Molinari, M., Calanca, V., Galli, C., Lucca, P. & Paganetti, P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299, 1397–1400 (2003).
Oda, Y., Hosokawa, N., Wada, I. & Nagata, K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299, 1394–1397 (2003).
Shenkman, M. et al. A shared endoplasmic reticulum-associated degradation pathway involving the EDEM1 protein for glycosylated and nonglycosylated proteins. J. Biol. Chem. 288, 2167–2178 (2013).
Tang, H. Y., Huang, C. H., Zhuang, Y. H., Christianson, J. C. & Chen, X. EDEM2 and OS-9 are required for ER-associated degradation of non-glycosylated sonic hedgehog. PLoS ONE 9, e92164 (2014).
Ushioda, R. et al. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569–572 (2008).
Hosokawa, N. et al. EDEM1 accelerates the trimming of α1,2-linked mannose on the C branch of N-glycans. Glycobiology 20, 567–575 (2010).
Olivari, S., Galli, C., Alanen, H., Ruddock, L. & Molinari, M. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 280, 2424–2428 (2005).
Mast, S. W. et al. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 15, 421–436 (2005).
Hirao, K. et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 281, 9650–9658 (2006).
Ninagawa, S. et al. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J. Cell Biol. 206, 347–356 (2014).
Hosokawa, N., Kamiya, Y., Kamiya, D., Kato, K. & Nagata, K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J. Biol. Chem. 284, 17061–17068 (2009).
Bernasconi, R., Pertel, T., Luban, J. & Molinari, M. A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: inhibiting secretion of misfolded protein conformers and enhancing their disposal. J. Biol. Chem. 283, 16446–16454 (2008).
Hosokawa, N. et al. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1–SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 283, 20914–20924 (2008).
Fujimori, T., Kamiya, Y., Nagata, K., Kato, K. & Hosokawa, N. Endoplasmic reticulum lectin XTP3-B inhibits endoplasmic reticulum-associated degradation of a misfolded α1-antitrypsin variant. FEBS J. 280, 1563–1575 (2013).
Tyler, R. E. et al. Unassembled CD147 is an endogenous endoplasmic reticulum-associated degradation substrate. Mol. Biol. Cell 23, 4668–4678 (2012).
Dersh, D., Jones, S. M., Eletto, D., Christianson, J. C. & Argon, Y. OS-9 facilitates turnover of nonnative GRP94 marked by hyperglycosylation. Mol. Biol. Cell 25, 2220–2234 (2014).
Bernasconi, R., Galli, C., Calanca, V., Nakajima, T. & Molinari, M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J. Cell Biol. 188, 223–235 (2010).
Alcock, F. & Swanton, E. Mammalian OS-9 is upregulated in response to endoplasmic reticulum stress and facilitates ubiquitination of misfolded glycoproteins. J. Mol. Biol. 385, 1032–1042 (2009).
Mikami, K. et al. The sugar-binding ability of human OS-9 and its involvement in ER-associated degradation. Glycobiology 20, 310–321 (2010).
Satoh, T. et al. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol. Cell 40, 905–916 (2010).
Travers, K. J. et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258 (2000).
Goder, V. & Melero, A. Protein O-mannosyltransferases participate in ER protein quality control. J. Cell Sci. 124, 144–153 (2011).
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W. & Prasher, D. C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
Aronson, D. E., Costantini, L. M. & Snapp, E. L. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic 12, 543–548 (2011).
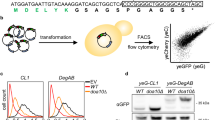
Xu, C., Wang, S., Thibault, G. & Ng, D. T. Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science 340, 978–981 (2013).
Denic, V., Quan, E. M. & Weissman, J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126, 349–359 (2006).
Acknowledgements
The authors wish to express their sincere apologies to those researchers whose work is not cited owing to the scope of the review and space constraints, and thank Kun Yang for assistance in rendering figures. Work in the authors' laboratories is supported by funds from the Temasek Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- HRD1 complex
-
Multi-subunit membrane protein complex in the endoplasmic reticulum (ER) that is organized around the E3 ubiquitin ligase HMG-CoA reductase degradation 1 (Hrd1). The HRD1 complex receives, retrotranslocates and ubiquitylates substrates for degradation by ER-associated degradation (ERAD).
- Lectin
-
Member of a class of proteins that bind to carbohydrates. Lectins usually have high specificity for sugar type and/or glycan-linkage configuration.
- Oxidoreductase
-
Member of a class of enzymes that mediate the transfer of electrons from one molecule to another. In the endoplasmic reticulum, most oxidoreductases form and break disulfide bonds.
- Proteoliposomes
-
Particles composed of proteins and lipids, which are usually reconstituted from purified components.
- DnaJ protein
-
A protein containing a 'J domain', which interacts with Hsp70 chaperones and stimulates their ATPase activity. Many DnaJ proteins are chaperones and directly bind to substrates.
- Unfolded protein response
-
(UPR). A signalling response that is triggered by the accumulation of misfolded or unfolded proteins in the endoplasmic reticulum.
- Microsomes
-
Vesicles that are formed from endoplasmic reticulum membranes after mechanical cell disruption.
Rights and permissions
About this article
Cite this article
Xu, C., Ng, D. Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol 16, 742–752 (2015). https://doi.org/10.1038/nrm4073
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm4073
This article is cited by
-
Genome-wide screen identifies new set of genes for improved heterologous laccase expression in Saccharomyces cerevisiae
Microbial Cell Factories (2024)
-
FAT4 overexpression promotes antitumor immunity by regulating the β-catenin/STT3/PD-L1 axis in cervical cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
Mutant p53-ENTPD5 control of the calnexin/calreticulin cycle: a druggable target for inhibiting integrin-α5-driven metastasis
Journal of Experimental & Clinical Cancer Research (2023)
-
USP2 promotes tumor immune evasion via deubiquitination and stabilization of PD-L1
Cell Death & Differentiation (2023)
-
Control of immune cell function by the unfolded protein response
Nature Reviews Immunology (2023)