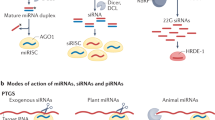
Key Points
-
Small RNAs can move between cells and function at transcriptional and post-transcriptional levels to transmit gene regulatory information.
-
In plants, both 24-nucleotide-long (24-nt-long) and 21-nt-long siRNA duplexes are able to move from cell to cell over short ranges and over long distances through the phloem. However, whether the small RNAs can transmit information may depend on the recipient cell.
-
In Caenorhabditis elegans, specific channel proteins are required for small RNA mobility, but their mechanisms are still unclear.
-
Mobile RNAs can be inherited from one generation to the next and can move from one organism to another, even crossing species barriers.
-
There is evidence that small RNAs are secreted from mammalian cells, but their functional importance is still unclear.
Abstract
Small RNAs that function in a non-cell autonomous manner are becoming increasingly recognized as regulatory molecules with the potential to transmit information between cells, organisms and species. In plants and nematodes, small RNA mobility can be genetically dissected to provide information about the nature of the mobile RNA species, their distribution in the organism and inside cells, as well as the cellular machinery required for mobility, including channel proteins and cellular trafficking factors. Mobile RNAs function in antiviral defence, cell signalling and gene expression regulation, and might also mediate transgenerational epigenetic inheritance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dunoyer, P. et al. Small RNA duplexes function as mobile silencing signals between plant cells. Science 328, 912–916 (2010). Shows for the first time that 21-nt-long siRNA duplexes are responsible for RNAi spreading over short distances in plants.
Molnar, A. et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875 (2010). Demonstrates an elegant combination of grafting and deep sequencing, which shows that 24-nt-long siRNAs are responsible for transmitting RNAi effects over long distances through the phloem.
Dunoyer, P. et al. An endogenous, systemic RNAi pathway in plants. EMBO J. 29, 1699–1712 (2010).
Melnyk, C. W., Molnar, A., Bassett, A. & Baulcombe, D. C. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Biol. 21, 1678–1683 (2011).
Winston, W. M., Molodowitch, C. & Hunter, C. P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 (2002). The first molecular characterization of a C. elegans protein that is specifically required for RNAi spreading.
Voinnet, O., Lederer, C. & Baulcombe, D. C. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103, 157–167 (2000). Demonstrates that viruses have evolved mechanisms to target mobile RNA, thus suggesting an important function for mobile RNAi in antiviral defence.
Carlsbecker, A. et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321 (2010). Demonstrates short-range mobility of miRNAs in plants and that it has a role in specifying development of the root.
Ashe, A. et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99 (2012).
Shirayama, M. et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77 (2012).
Buckley, B. A. et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451 (2012). Shows, together with references 8 and 9, that piRNAs and RNAi induce transgenerational epigenetic silencing in C. elegans.
Weiberg, A. et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013). Reports the transfer of small RNAs between species.
Palauqui, J. C., Elmayan, T., Pollien, J. M. & Vaucheret, H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745 (1997).
Voinnet, O., Vain, P., Angell, S. & Baulcombe, D. C. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187 (1998).
Voinnet, O. & Baulcombe, D. C. Systemic signalling in gene silencing. Nature 389, 553 (1997).
Hamilton, A., Voinnet, O., Chappell, L. & Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 (2002).
Brosnan, C. A. & Voinnet, O. Cell-to-cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr. Opin. Plant Biol. 14, 580–587 (2011).
Dunoyer, P., Himber, C., Ruiz-Ferrer, V., Alioua, A. & Voinnet, O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nature Genet. 39, 848–856 (2007).
Smith, L. M. et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19, 1507–1521 (2007).
Buhtz, A., Springer, F., Chappell, L., Baulcombe, D. C. & Kehr, J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 53, 739–749 (2008).
Varkonyi-Gasic, E., Gould, N., Sandanayaka, M., Sutherland, P. & MacDiarmid, R. M. Characterisation of microRNAs from apple (Malus domestica 'Royal Gala') vascular tissue and phloem sap. BMC Plant Biol. 10, 159 (2010).
Molnar, A., Melnyk, C. & Baulcombe, D. C. Silencing signals in plants: a long journey for small RNAs. Genome Biol. 12, 215 (2011).
Melnyk, C. W., Molnar, A. & Baulcombe, D. C. Intercellular and systemic movement of RNA silencing signals. EMBO J. 30, 3553–3563 (2011).
Zhou, G.-K., Kubo, M., Zhong, R., Demura, T. & Ye, Z.-H. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 48, 391–404 (2007).
Miyashima, S., Koi, S., Hashimoto, T. & Nakajima, K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313 (2011).
Pant, B. D., Buhtz, A., Kehr, J. & Scheible, W.-R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738 (2008).
Lin, S.-I. et al. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732–746 (2008).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). The first demonstration of RNAi in C. elegans , which already provides clear evidence that RNAi systemically spreads throughout the organism.
Timmons, L. & Fire, A. Specific interference by ingested dsRNA. Nature 395, 854 (1998). The discovery that C. elegans can ingest dsRNA and process it to silence endogenous genes.
Timmons, L., Court, D. L. & Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 (2001).
Hinas, A., Wright, A. J. & Hunter, C. P. SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans. Curr. Biol. 22, 1938–1943 (2012).
Winston, W. M., Sutherlin, M., Wright, A. J., Feinberg, E. H. & Hunter, C. P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl Acad. Sci. USA 104, 10565–10570 (2007). Identifies the channel protein responsible for the uptake of dsRNA from the environment by C. elegans.
Jose, A. M., Kim, Y. A., Leal-Ekman, S. & Hunter, C. P. Conserved tyrosine kinase promotes the import of silencing RNA into Caenorhabditis elegans cells. Proc. Natl Acad. Sci. USA 109, 14520–14525 (2012).
Feinberg, E. H. & Hunter, C. P. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301, 1545–1547 (2003).
Shih, J. D., Fitzgerald, M. C., Sutherlin, M. & Hunter, C. P. The SID-1 double-stranded RNA transporter is not selective for dsRNA length. RNA 15, 384–390 (2009).
Saleh, M.-C. et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nature Cell Biol. 8, 793–802 (2006).
Jose, A. M., Garcia, G. A. & Hunter, C. P. Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Nature Struct. Mol. Biol. 18, 1184–1188 (2011).
Parrish, S. & Fire, A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7, 1397–1402 (2001).
Tabara, H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999).
Pak, J., Maniar, J. M., Mello, C. C. & Fire, A. Protection from feed-forward amplification in an amplified RNAi mechanism. Cell 151, 885–899 (2012).
Aphasizhev, R. RNA uridylyltransferases. Cell. Mol. Life Sci. 62, 2194–2203 (2005).
McEwan, D. L., Weisman, A. S. & Hunter, C. P. Uptake of extracellular double-stranded RNA by SID-2. Mol. Cell 47, 746–754 (2012).
Sundaram, P., Echalier, B., Han, W., Hull, D. & Timmons, L. ATP-binding cassette transporters are required for efficient RNA interference in Caenorhabditis elegans. Mol. Biol. Cel. 17, 3678–3688 (2006).
Nuez, I. & Félix, M.-A. Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS ONE 7, e29811 (2012).
Ibrahim, H. M. M. et al. Post-transcriptional gene silencing of root-knot nematode in transformed soybean roots. Exp. Parasitol. 127, 90–99 (2011).
Maule, A. G. et al. An eye on RNAi in nematode parasites. Trends Parasitol. 27, 505–513 (2011).
Baulcombe, D. RNA silencing in plants. Nature 431, 356–363 (2004).
Brodersen, P. & Voinnet, O. The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280 (2006).
Bologna, N. G. & Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65, 473–503 (2014).
Havelda, Z., Hornyik, C., Crescenzi, A. & Burgyán, J. In situ characterization of Cymbidium Ringspot Tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 77, 6082–6086 (2003).
Deleris, A. et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71 (2006).
Guo, H. S. & Ding, S. W. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21, 398–407 (2002).
Hamera, S., Song, X., Su, L., Chen, X. & Fang, R. Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 69, 104–115 (2012).
Duan, C.-G. et al. Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 24, 259–274 (2012).
Lu, R. et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436, 1040–1043 (2005).
Li, H., Li, W. X. & Ding, S. W. Induction and suppression of RNA silencing by an animal virus. Science 296, 1319–1321 (2002).
Wilkins, C. et al. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 436, 1044–1047 (2005).
Schott, D. H., Cureton, D. K., Whelan, S. P. & Hunter, C. P. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 102, 18420–18424 (2005).
Félix, M.-A. et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9, e1000586 (2011).
Ashe, A. et al. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. eLife 2, e00994 (2013).
Duchaine, T. F. et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124, 343–354 (2006).
Lu, R., Yigit, E., Li, W. X. & Ding, S. W. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 5, e1000286 (2009).
Rehwinkel, J. & Reis e Sousa, C. RIGorous detection: exposing virus through RNA sensing. Science 327, 284–286 (2010).
Corrêa, R. L., Steiner, F. A., Berezikov, E. & Ketting, R. F. MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 6, e1000903 (2010).
Sarkies, P., Ashe, A., Le Pen, J., McKie, M. A. & Miska, E. A. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 23, 1258–1270 (2013).
Shivaprasad, P. V. et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874 (2012).
Marín-González, E. & Suárez-López, P. “And yet it moves”: cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci. 196, 18–30 (2012).
Lee, H.-C. et al. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87 (2012).
Seth, M. et al. The C. elegans CSR-1 AGO pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell 27, 656–663 (2013).
Wedeles, C. J., Wu, M. Z. & Claycomb, J. M. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell 27, 664–671 (2013).
Tijsterman, M., May, R. C., Simmer, F., Okihara, K. L. & Plasterk, R. H. A. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr. Biol. 14, 111–116 (2004).
Zhang, C. et al. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr. Biol. 22, 881–890 (2012).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Zhang, H. & Fire, A. Z. Cell autonomous specification of temporal identity by Caenorhabditis elegans microRNA lin-4. Dev. Biol. 344, 603–610 (2010).
Heard, E. & Martienssen, R. A. Transgenerational Epigenetic Inheritance: Myths and Mechanisms. Cell 157, 95–109 (2014).
Law, J. A. & Jacobsen, S. E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Rev. Genet. 11, 204–220 (2010).
Gu, S. G. et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nature Genet. 44, 157–164 (2012).
Slotkin, R. K. et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472 (2009). Shows that mobile siRNAs in plants induce transposon silencing in the next generation.
Calarco, J. P. et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151, 194–205 (2012).
Ibarra, C. A. et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364 (2012).
Henderson, I. R. & Jacobsen, S. E. Epigenetic inheritance in plants. Nature 447, 418–424 (2007).
Grant-Downton, R. et al. Artificial microRNAs reveal cell-specific differences in small RNA activity in pollen. Curr. Biol. 23, R599–R601 (2013).
Siomi, M. C., Sato, K., Pezic, D. & Aravin, A. A. PIWI-interacting small RNAs: the vanguard of genome defence. Nature Rev. Mol. Cell Biol. 12, 246–258 (2011).
Das, P. P. et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31, 79–90 (2008).
Batista, P. J. et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78 (2008).
Bagijn, M. P. et al. Function, targets, and Eevolution of Caenorhabditis elegans piRNAs. Science 337, 574–578 (2012).
Luteijn, M. J. et al. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31, 3422–3430 (2012).
Alcazar, R. M., Lin, R. & Fire, A. Z. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics 180, 1275–1288 (2008).
Zhuang, J. J., Banse, S. A. & Hunter, C. P. The nuclear argonaute NRDE-3 contributes to transitive RNAi in Caenorhabditis elegans. Genetics 194, 117–131 (2013).
Sarkies, P. & Miska, E. A. RNAi pathways in the recognition of foreign RNA: antiviral responses and host-parasite interactions in nematodes. Biochem. Soc. Trans. 41, 876–880 (2013).
Sarkies, P. & Miska, E. A. Molecular biology. Is there social RNA? Science 341, 467–468 (2013).
Zong, J., Yao, X., Yin, J., Zhang, D. & Ma, H. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: Duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 447, 29–39 (2009).
Turchinovich, A., Weiz, L. & Burwinkel, B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 37, 460–465 (2012).
Gibbings, D. J., Ciaudo, C., Erhardt, M. & Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nature Cell Biol. 11, 1143–1149 (2009).
Lee, Y. S. et al. Silencing by small RNAs is linked to endosomal trafficking. Nature Cell Biol. 11, 1150–1156 (2009).
Gibbings, D. & Voinnet, O. Control of RNA silencing and localization by endolysosomes. Trends Cell Biol. 20, 491–501 (2010).
Yao, B., La, L. B., Chen, Y.-C., Chang, L.-J. & Chan, E. K. L. Defining a new role of GW182 in maintaining miRNA stability. EMBO Rep. 13, 1102–1108 (2012).
Li, Y., Lu, J., Han, Y., Fan, X. & Ding, S. W. RNA interference functions as an antiviral immunity mechanism in mammals. Science 342, 231–234 (2013).
Maillard, P. V. et al. Antiviral RNA interference in mammalian cells. Science 342, 235–238 (2013).
Shapiro, E., Biezuner, T. & Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature Rev. Genet. 14, 618–630 (2013).
Okamura, K. & Lai, E. C. Endogenous small interfering RNAs in animals. Nature Rev. Mol. Cell Biol. 9, 673–678 (2008).
Hutvagner, G. & Simard, M. J. Argonaute proteins: key players in RNA silencing. Nature Rev. Mol. Cell Biol. 9, 22–32 (2008).
Pak, J. & Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241–244 (2007).
Olovnikov, I., Aravin, A. A. & Toth, K. F. Small RNA in the nucleus: the RNA-chromatin ping-pong. Curr. Opin. Genet. Dev. 22, 164–171 (2012).
Henderson, I. R. et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nature Genet. 38, 721–725 (2006).
Xie, Z. et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, E104 (2004).
Zilberman, D. et al. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 14, 1214–1220 (2004).
Kasschau, K. D. et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5, e57 (2007).
Papp, I. et al. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132, 1382–1390 (2003).
Knight, S. W. & Bass, B. L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269–2271 (2001).
Ketting, R. F. et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659 (2001).
Sijen, T., Steiner, F. A., Thijssen, K. L. & Plasterk, R. H. A. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247 (2007).
Fischer, S. E. J. et al. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 7, e1002369 (2011).
Gent, J. I. et al. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 37, 679–689 (2010).
Jablonka, E. & Lamb, M. J. Soft inheritance: challenging the modern synthesis. Genet. Mol. Biol. 31, 2 (2008).
Luna, E., Bruce, T. J. A., Roberts, M. R., Flors, V. & Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853 (2012).
Dowen, R. H. et al. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl Acad. Sci. USA 109, E2183–2191 (2012).
Bond, D. M. & Baulcombe, D. C. Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 24, 100–107 (2014).
Acknowledgements
The authors thank members of the Miska laboratory for helpful discussions. P.S. was funded by a Gonville and Caius College Research Fellowship. Work in the Miska laboratory was funded by Cancer Research UK and the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (box)
Antiviral RNAi in mammals (PDF 232 kb)
Glossary
- microRNAs
-
(miRNAs). RNAs that are ∼20–24 nucleotides in length and derived from specific endogenous loci. They are transcribed to form hairpin precursors that are cleaved to result in the final mature miRNA and miRNA* (corresponding to the more and less abundant RNAs, respectively) sequences.
- Grafting
-
A technique widely used in agriculture and plant research that enables the stem of one plant to be joined to the roots of another. Vessels such as the xylem and the phloem fuse to enable the transfer of material between the two parts of the plant, enabling the grafted plant to survive and grow healthily.
- Scion
-
A detached portion of a plant (for example, a bud or a shoot) joined to a stock in grafting experiments and usually only providing aerial parts to a graft.
- Co-suppression
-
Discovered in plants, co-suppression occurs when a transgene can cause silencing of the endogenous copies of the homologous gene. Co-suppression is caused by small RNAs made against the transgene that can function in trans to silence endogenous loci.
- Phloem
-
In vascular plants, the phloem is the main nutrient transport system, which carries organic nutrients (mostly sucrose- containing fluids) from tissues where they are synthesized ('source') to other tissues where they are needed ('sink').
- Agrobacterium tumefaciens
-
A soil-dwelling bacterium that naturally infects plants to form crown gall tumours by inserting an invading DNA sequence (called T-DNA) into the plant genome. In the laboratory, engineered versions of A. tumefaciens can be used to insert transgenes of interest into plant cells.
- Plasmodesmata
-
Large channels between adjacent plant cells that are directly formed by fusion of the cell membranes. They traverse the cell walls, to enable flow between the cytoplasm, and are typically around 50 nm in diameter (large enough to enable protein traffic).
- Dicer enzyme
-
These enzymes are members of the class II subgroup of ribonuclease III enzymes that are found across Eukarya and function to generate both microRNAs and siRNAs through cleavage of precursors containing double-stranded regions. They are known as Dicer-like enzymes in plants.
- Argonaute protein
-
(AGO protein). A member of a family of binding partners and effectors for many types of small RNA including those generated by Dicer but also PIWI-interacting RNAs (piRNAs) and other Dicer- independent small RNAs. Typically, AGO proteins, in conjunction with accessory proteins and their small RNA cofactors, form RNA-induced silencing complexes to bring about target gene silencing.
- Sieve cells
-
Specialized cells lacking a nucleus that enable the flow through the phloem between enlarged plasmodesmata.
- GW182
-
An accessory protein for Argonaute-silencing proteins, characterized by multiple Trp-Gly repeats.
- ABC transporter
-
A widely conserved group of transmembrane proteins that power the transport of a range of different substrates through the hydrolysis of ATP.
- Suppressors of silencing
-
Virally encoded proteins that enable viruses to counter RNAi defence mechanisms. Most examples of these proteins known so far derive from plant viruses and inhibit RNAi at different stages in the pathway.
- Positive-strand RNA virus
-
The RNA genome of this virus encodes the sense strand of the viral proteins, thus it can be directly translated to produce a coding sequence. This is in contrast to negative-strand viruses, which must be copied to produce the positive strand before translation can occur.
- Interferon response
-
A key component of the mammalian innate immune pathway. It involves transcriptional activation of interferon genes, which in turn induce intracellular and intercellular effects.
- Vegetative nucleus
-
One of the two nuclei in the plant pollen. It does not contribute DNA to the next generation but instead forms the endosperm after fertilization, which provides nourishment to the developing seed.
Rights and permissions
About this article
Cite this article
Sarkies, P., Miska, E. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol 15, 525–535 (2014). https://doi.org/10.1038/nrm3840
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3840
This article is cited by
-
Genome-wide identification, characterization and expression analysis of AGO, DCL, and RDR families in Chenopodium quinoa
Scientific Reports (2023)
-
Soma-to-germline RNA communication
Nature Reviews Genetics (2022)
-
Rethinking hereditary relations: the reconstitutor as the evolutionary unit of heredity
Synthese (2022)
-
The effect of miR-539 regulating TRIAP1 on the apoptosis, proliferation, migration and invasion of osteosarcoma cells
Cancer Cell International (2021)
-
Examining the evidence for extracellular RNA function in mammals
Nature Reviews Genetics (2021)