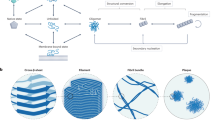
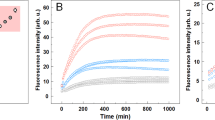
Key Points
-
The phenomenon of amyloid formation is associated with protein misfolding disorders, including Alzheimer's disease, Parkinson's disease and type II diabetes.
-
The amyloid state is a 'generic state' of proteins and its study can provide great insight into the nature of functional structures and into that of disease-related assemblies.
-
A multitude of quality control, or 'housekeeping', mechanisms exist in living organisms to prevent the conversion of normally soluble proteins into the aberrant amyloid state and to maintain protein homeostasis.
-
The failure of these quality control mechanisms can give rise to 'protein metastasis', the uncontrolled conversion of these molecules into aberrant self-propagating assemblies that ultimately lead to a cascade of cytotoxic processes.
-
Our increasing ability to monitor and characterize the molecular structures and formation mechanisms of the protein species that are involved in amyloid formation is suggesting novel strategies to treat or prevent protein misfolding disorders.
-
Ultimately, the results of this field of research will result in great changes in the way we are able to manage modern lifestyles and maintain healthy ageing.
Abstract
The phenomenon of protein aggregation and amyloid formation has become the subject of rapidly increasing research activities across a wide range of scientific disciplines. Such activities have been stimulated by the association of amyloid deposition with a range of debilitating medical disorders, from Alzheimer's disease to type II diabetes, many of which are major threats to human health and welfare in the modern world. It has become clear, however, that the ability to form the amyloid state is more general than previously imagined, and that its study can provide unique insights into the nature of the functional forms of peptides and proteins, as well as understanding the means by which protein homeostasis can be maintained and protein metastasis avoided.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
12 June 2014
In the legend of figure 2 of the above article (page 388), the sentence “The spacing between polypeptide chains along the fibril axis is constant to a good approximation even for very different polypeptide sequences, a generic property arising from the common inter-side chain hydrogen bonding constraints (orange line in part b).” incorrectly referred to 'inter-side chain' hydrogen bonding constraints, whereas it should have referred to 'inter-main chain' hydrogen bonding constraints. This has been corrected online. The authors apologize for any confusion caused to readers.
References
Dobson, C. M. Protein folding and misfolding. Nature 426, 884–890 (2003).
Hardy, J. & Selkoe, D. J. Medicine - the amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002).
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008). Provides a comprehensive overview of protein homeostasis and of the opportunities for therapeutic intervention that it offers.
Tanaka, M., Collins, S. R., Toyama, B. H. & Weissman, J. S. The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 (2006).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Bioch. 75, 333–366 (2006).
Knowles, T. P. J. & Buehler, M. J. Nanomechanics of functional and pathological amyloid materials. Nature Nanotech. 6, 469–479 (2011).
Haass, C. & Selkoe, D. J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nature Rev. Mol. Cell. Biol. 8, 101–112 (2007).
Eisenberg, D. & Jucker, M. The amyloid state of proteins in human diseases. Cell 148, 1188–1203 (2012).
Ballard, C. et al. Alzheimer's disease. Lancet 377, 1019–1031 (2011).
Querfurth, H. W. & LaFerla, F. M. Mechanisms of disease Alzheimer's disease. N. Engl. J. Med. 362, 329–344 (2010).
Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M. & Ulrich Hartl, F. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Bioch. 82, 323–355 (2013).
Alzheimer's disease international. World Alzheimer report (2010).
Dementia: A public health priority. World health organization and Alzheimer's disease international (2012).
Olshansky, S. J. et al. A potential decline in life expectancy in the united states in the 21st century. N. Engl. J. Med. 352, 1138–1145 (2005).
Alzheimer's disease: Facts and figures. Alzheimer's association (2012).
Walsh, D. M. et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 (2002).
Cremades, N. et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell 149, 1048–1059 (2012).
Cohen, S. I. A. et al. Proliferation of amyloid-β 42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl Acad. Sci. USA 110, 9758–9763 (2013). Shows how the rapid proliferation of amyloid aggregates can be catalysed by the surfaces of existing amyloid fibrils.
Lesne, S. et al. A specific amyloid-β protein assembly in the brain impairs memory. Nature 440, 352–357 (2006).
Campioni, S. et al. A causative link between the structure of aberrant protein oligomers and their toxicity. Nature Chem. Biol. 6, 140–147 (2010).
Bucciantini, M. et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 (2002).
Caughey, B. & Lansbury, P. T. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26, 267–298 (2003).
Billings, L. M., Oddo, S., Green, K. N., McGaugh, J. L. & LaFerla, F. M. Intraneuronal a β causes the onset of early Alzheimer's disease-related cognitive deficits in transgenic mice. Neuron 45, 675–688 (2005).
Winner, B. et al. In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl Acad. Sci. USA 108, 4194–4199 (2011).
Koffie, R. M. et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl Acad. Sci. USA 106, 4012–4017 (2009).
Dobson, C. M. Protein misfolding, evolution and disease. Trends Bioch. Sci. 24, 329–332 (1999).
Greenwald, J. & Riek, R. On the possible amyloid origin of protein folds. J. Mol. Biol. 421, 417–426 (2012).
Carny, O. & Gazit, E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 19, 1051–1055 (2005).
DePas, W. H. & Chapman, M. R. Microbial manipulation of the amyloid fold. Res. Microbiol. 163, 592–606 (2012).
Fowler, D. M., Koulov, A. V., Balch, W. E. & Kelly, J. W. Functional amyloid - from bacteria to humans. Trends Bioch. Sci. 32, 217–224 (2007).
Fandrich, M. & Dobson, C. M. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 21, 5682–5690 (2002).
Knowles, T. P. J. et al. Observation of spatial propagation of amyloid assembly from single nuclei. Proc. Natl Acad. Sci. USA 108, 14746–14751 (2011).
Astbury, W. T., Dickinson, S. & Bailey, K. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J. 29, 2351–2360 (1935).
Kendrew, J. C. et al. 3-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature 181, 662–666 (1958).
Berman, H. M. et al. The protein data bank. Nucl. Acids Res. 28, 235–242 (2000).
Rosenbaum, D. M., Rasmussen, S. G. F. & Kobilka, B. K. The structure and function of g-protein-coupled receptors. Nature 459, 356–363 (2009).
Anfinsen, C. B. Principles that govern folding of protein chains. Science 181, 223–230 (1973).
Dill, K. A. & Chan, H. S. From Levinthal to pathways to funnels. Nature Struct. Biol. 4, 10–19 (1997).
Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997).
Dobson, C. M., Sali, A. & Karplus, M. Protein folding: a perspective from theory and experiment. Angew. Chem. Int. Ed. 37, 868–893 (1998).
Baldwin, A. J. et al. Metastability of native proteins and the phenomenon of amyloid formation. J. Am. Chem. Soc. 133, 14160–14163 (2011). Demonstrates that native states of proteins are intrinsically metastable against aggregation.
Gazit, E. The “correctly folded” state of proteins: is it a metastable state. Angew. Chem. Int. Ed. 41, 257–259 (2002).
Vendruscolo, M., Knowles, T. P. J. & Dobson, C. M. Protein solubility and protein homeostasis: A generic view of protein misfolding disorders. Cold Spring Harb. Perspect. Biol. 3, a010454 (2011).
Wolff, S., Weissman, J. S. & Dillin, A. Differential scales of protein quality control. Cell 157, 52–64 (2014).
Dyson, H. J. & Wright, P. E. Intrinsically unstructured proteins and their functions. Nature Rev. Mol. Cell Biol. 6, 197–208 (2005).
Uversky, V. N. & Dunker, A. K. Understanding protein non-folding. Biochim. Biophys. Acta 1804, 1231–1264 (2010).
Dedmon, M. M., Lindorff-Larsen, K., Christodoulou, J., Vendruscolo, M. & Dobson, C. M. Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 127, 476–477 (2005).
Hou, L. M. et al. Solution NMR studies of the Aβ(1-40) and Aβ(1-42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J. Am. Chem. Soc. 126, 1992–2005 (2004).
Varadi, M. et al. pE-DB: A database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucl. Acids Res. 42, D326–D335 (2014).
Tartaglia, G. G. et al. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 380, 425–436 (2008).
Polymeropoulos, M. H. et al. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science 276, 2045–2047 (1997).
Spillantini, M. G. et al. α-synuclein in Lewy bodies. Nature 388, 839–840 (1997).
Westermark, P. et al. Amyloid fibrils in human insulinoma and islets of langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc. Natl Acad. Sci. USA 84, 3881–3885 (1987).
Waudby, C. A., Launay, H., Cabrita, L. D. & Christodoulou, J. Protein folding on the ribosome studied using NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 74, 57–75 (2013).
Chiti, F. & Dobson, C. M. Amyloid formation by globular proteins under native conditions. Nature Chem. Biol. 5, 15–22 (2009).
Kelly, J. W. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 8, 101–106 (1998).
Vendruscolo, M. & Dobson, C. M. Structural biology: protein self-assembly intermediates. Nature Chem. Biol. 9, 216–217 (2013).
Neupert, W. & Herrmann, J. M. Translocation of proteins into mitochondria. Annu. Rev. Bioch. 76, 723–749 (2007).
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Meinema, A. C. et al. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science 333, 90–93 (2011).
Park, S. H. et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154, 134–145 (2013).
Fletcher, D. A. & Mullins, D. Cell mechanics and the cytoskeleton. Nature 463, 485–492 (2010).
Omenetto, F. G. & Kaplan, D. L. New opportunities for an ancient material. Science 329, 528–531 (2010).
Anderson, V. J. & Lekkerkerker, H. N. W. Insights into phase transition kinetics from colloid science. Nature 416, 811–815 (2002).
Prusiner, S. B. Prions. Proc. Natl Acad. Sci. USA 95, 13363–13383 (1998).
Serio, T. R. et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 (2000).
Sipe, J. D. & Cohen, A. S. Review: History of the amyloid fibril. J. Struct. Biol. 130, 88–98 (2000).
Buxbaum, J. N. & Linke, R. P. A molecular history of the amyloidoses. J. Mol. Biol. 421, 142–159 (2012).
Stefani, M. & Dobson, C. M. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. 81, 678–699 (2003).
Liao, L. et al. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J. Biol. Chem. 279, 37061–37068 (2004).
Wang, Q. et al. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB J. 19, 869–871 (2005).
Xia, Q. et al. Proteomic identification of novel proteins associated with lewy bodies. Front. Biosci. 13, 3850–3856 (2008).
Sunde, M. et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273, 729–739 (1997).
Sawaya, M. R. et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447, 453–457 (2007).
Fitzpatrick, A. W. P. et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc. Natl Acad. Sci. USA 110, 5468–5473 (2013).
Petkova, A. T. et al. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl Acad. Sci. USA 99, 16742–16747 (2002).
Wasmer, C. et al. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science 319, 1523–1526 (2008).
Luhrs, T. et al. 3D structure of Alzheimer's amyloid-β(1-42) fibrils. Proc. Natl Acad. Sci. USA 102, 17342–17347 (2005).
Guijarro, J. I., Sunde, M., Jones, J. A., Campbell, I. D. & Dobson, C. M. Amyloid fibril formation by an SH3 domain. Proc. Natl Acad. Sci. USA 95, 4224–4228 (1998).
Chiti, F. et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl Acad. Sci. USA 96, 3590–3594 (1999).
Urbanc, B. et al. In silico study of amyloid β-protein folding and oligomerization. Proc. Natl Acad. Sci. USA 101, 17345–17350 (2004).
Auer, S., Meersman, F., Dobson, C. M. & Vendruscolo, M. A generic mechanism of emergence of amyloid protofilaments from disordered oligomeric aggregates. PLoS Comp. Biol. 4, e1000222 (2008).
Tycko, R. Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 62, 279–299 (2011).
Jimenez, J. L. et al. The protofilament structure of insulin amyloid fibrils. Proc. Natl Acad. Sci. USA 99, 9196–9201 (2002).
Sachse, C., Fandrich, M. & Grigorieff, N. Paired beta-sheet structure of an Aβ(1-40) amyloid fibril revealed by electron microscopy. Proc. Natl Acad. Sci. USA 105, 7462–7466 (2008).
Knowles, T. P. et al. Role of intermolecular forces in defining material properties of protein nanofibrils. Science 318, 1900–1903 (2007).
Krishnan, R. & Lindquist, S. L. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005).
Collinge, J. & Clarke, A. R. A general model of prion strains and their pathogenicity. Science 318, 930–936 (2007).
Halfmann, R., Alberti, S. & Lindquist, S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 20, 125–133 (2010).
Wright, C. F., Teichmann, S. A., Clarke, J. & Dobson, C. M. The importance of sequence diversity in the aggregation and evolution of proteins. Nature 438, 878–881 (2005).
Knowles, T. P. J. et al. Twisting transition between crystalline and fibrillar phases of aggregated peptides. Phys. Rev. Lett. 109, 158101 (2012).
Smith, J. F., Knowles, T. P. J., Dobson, C. M., MacPhee, C. E. & Welland, M. E. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl Acad. Sci. USA 103, 15806–15811 (2006).
Tartaglia, G. G., Pechmann, S., Dobson, C. M. & Vendruscolo, M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Bioch. Sci. 32, 204–206 (2007).
Apetri, M. M., Maiti, N. C., Zagorski, M. G., Carey, P. R. & Anderson, V. E. Secondary structure of α -synuclein oligomers: characterization by Raman and atomic force microscopy. J. Mol. Biol. 355, 63–71 (2006).
Nettleton, E. J. et al. Characterization of the oligomeric states of insulin in self-assembly and amyloid fibril formation by mass spectrometry. Bioph. J. 79, 1053–1065 (2000).
Smith, D. P., Radford, S. E. & Ashcroft, A. E. Elongated oligomers in β2-microglobulin amyloid assembly revealed by ion mobility spectrometry-mass spectrometry. Proc. Natl Acad. Sci. USA 107, 6794–6798 (2010).
Bernstein, S. L. et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nature Chem. 1, 326–331 (2009).
Narayan, P. et al. The extracellular chaperone clusterin sequesters oligomeric forms of the amyloid-β1–40 peptide. Nature Struct. Mol. Biol. 19, 79–83 (2012).
Cohen, S. I. A., Vendruscolo, M., Dobson, C. M. & Knowles, T. P. J. From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. 421, 160–171 (2012).
Knowles, T. P. J. et al. An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009). Presents an analytical solution to the kinetic equations that describe protein aggregation, thus providing an effective method to identify the roles of the individual microscopic processes underlying protein aggregation.
ten Wolde, P.R. & Frenkel, D. Enhancement of protein crystal nucleation by critical density fluctuations. Science 277, 1975–1978 (1997).
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Jucker, M. & Walker, L. C. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann. Neurol. 70, 532–540 (2011).
Fandrich, M. Absolute correlation between lag time and growth rate in the spontaneous formation of several amyloid-like aggregates and fibrils. J. Mol. Biol. 365, 1266–1270 (2007).
Collins, S. R., Douglass, A., Vale, R. D. & Weissman, J. S. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2, 1582–1590 (2004).
Bolognesi, B. et al. Single point mutations induce a switch in the molecular mechanism of the aggregation of the Alzheimer's disease associated Aβ42 peptide. ACS Chem. Biol. 9, 378–382 (2013).
Meier, M. et al. Plug-based microfluidics with defined surface chemistry to miniaturize and control aggregation of amyloidogenic peptides. Angew. Chem. Int. Ed. 121, 1515–1517 (2009).
Lee, S. J., Desplats, P., Sigurdson, C., Tsigelny, I. & Masliah, E. Cell-to-cell transmission of non-prion protein aggregates. Nature Rev. Neurol. 6, 702–706 (2010).
Polymenidou, M. & Cleveland, D. W. The seeds of neurodegeneration: prion-like spreading in als. Cell 147, 498–508 (2011).
Cohen, S. et al. Spatial propagation of protein polymerization. Phys. Rev. Lett. 112, 098101 (2014).
Aguzzi, A. Cell biology: Beyond the prion principle. Nature 459, 924–925 (2009).
Aguzzi, A. & Rajendran, L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 64, 783–790 (2009).
Jucker, M. & Walker, L. C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 (2013).
Walker, L. C., Diamond, M. I., Duff, K. E. & Hyman, B. T. Mechanisms of protein seeding in neurodegenerative diseases. JAMA Neurol. 70, 304–310 (2013).
Maji, S. K. et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 (2009).
Chiti, F., Stefani, M., Taddei, N., Ramponi, G. & Dobson, C. M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 424, 805–808 (2003).
Dubay, K. F. et al. Prediction of the absolute aggregation rates of amyloidogenic polypeptide chains. J. Mol. Biol. 341, 1317–1326 (2004).
Fernandez-Escamilla, A. M., Rousseau, F., Schymkowitz, J. & Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nature Biotech. 22, 1302–1306 (2004).
Pawar, A. P. et al. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J. Mol. Biol. 350, 379–392 (2005).
Neudecker, P. et al. Structure of an intermediate state in protein folding and aggregation. Science 336, 362–366 (2012).
Broome, B. M. & Hecht, M. H. Nature disfavors sequences of alternating polar and non-polar amino acids: Implications for amyloidogenesis. J. Mol. Biol. 296, 961–968 (2000).
Richardson, J. S. & Richardson, D. C. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl Acad. Sci. USA 99, 2754–2759 (2002).
Otzen, D. E. & Oliveberg, M. Salt-induced detour through compact regions of the protein folding landscape. Proc. Natl Acad. Sci. USA 96, 11746–11751 (1999).
Kaganovich, D., Kopito, R. & Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 (2008).
Tyedmers, J., Mogk, A. & Bukau, B. Cellular strategies for controlling protein aggregation. Nature Rev. Mol. Cell. Biol. 11, 777–788 (2010).
Bence, N. F., Sampat, R. M. & Kopito, R. R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552–1555 (2001).
Ross, C. A. & Poirier, M. A. Protein aggregation and neurodegenerative disease. Nature Med. 10, S10–S17 (2004).
Glickman, M. H. & Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 (2002).
Rubinsztein, D. C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 (2006).
Webb, J. L., Ravikumar, B., Atkins, J., Skepper, J. N. & Rubinsztein, D. C. α-synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278, 25009–25013 (2003).
Mizushima, N., Levine, B., Cuervo, A. M. & Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Morimoto, R. I. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12, 3788–3796 (1998).
Jiang, Q. et al. ApoE promotes the proteolytic degradation of Aβ. Neuron 58, 681–693 (2008).
Knowles, T. P. J. et al. Kinetics and thermodynamics of amyloid formation from direct measurements of fluctuations in fibril mass. Proc. Natl Acad. Sci. USA 104, 10016–10021 (2007).
Xu, L. Q. et al. Influence of specific Hsp70 domains on fibril formation of the yeast prion protein Ure2. Philos. Trans. R. Soc. B 368, 20110410 (2013).
Wilson, M. R. & Easterbrook-Smith, S. B. Clusterin is a secreted mammalian chaperone. Trends Bioch. Sci. 25, 95–98 (2000).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature Genet. 41, 1088–1093 (2009).
Lambert, J.-C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature Genet. 45, 1452–1458 (2013).
Lambert, J. C. et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nature Genet. 41, 1094–1099 (2009).
Tomic, J. L., Pensalfini, A., Head, E. & Glabe, C. G. Soluble fibrillar oligomer levels are elevated in Alzheimer's disease brain and correlate with cognitive dysfunction. Neurobiol. Dis. 35, 352–358 (2009).
Shankar, G. M. et al. Natural oligomers of the Alzheimer amyloid-β protein induce reversible synapse loss by modulating an NMDA-type glutamate receptor-dependent signaling pathway. J. Neurosci. 27, 2866–2875 (2007).
Palop, J. J. & Mucke, L. Amyloid-β-induced neuronal dysfunction in Alzheimer's disease: from synapses toward neural networks. Nature Neurosci. 13, 812–818 (2010).
Feany, M. B. & Bender, W. W. A Drosophila model of Parkinson's disease. Nature 404, 394–398 (2000).
Luheshi, L. M., Crowther, D. C. & Dobson, C. M. Protein misfolding and disease: from the test tube to the organism. Curr. Opin. Chem. Biol. 12, 25–31 (2008).
Bilen, J. & Bonini, N. M. Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 39, 153–171 (2005).
Ben-Zvi, A., Miller, E. A. & Morimoto, R. I. Collapse of proteostasis represents an early molecular event in caenorhabditis elegans aging. Proc. Natl Acad. Sci. USA 106, 14914–14919 (2009).
Kaminksi Schierle, G. S. et al. In situ measurements of the formation and morphology of intracellular β-amyloid fibrils by super-resolution fluorescence imaging. J. Am. Chem. Soc. 133, 12902–12905 (2011).
McGuire, E. K. et al. Selenium-enhanced electron microscopic imaging of different aggregate forms of a segment of the amyloid beta peptide in cells. ACS Nano 6, 4740–4747 (2012).
Ries, J. et al. Superresolution imaging of amyloid fibrils with binding-activated probes. ACS Chem. Neurosci. 4, 1057–1061 (2013).
Lansbury, P. T. & Lashuel, H. A. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature 443, 774–779 (2006).
Karran, E., Mercken, M. & De Strooper, B. The amyloid cascade hypothesis for Alzheimer's disease: an appraisal for the development of therapeutics. Nature Rev. Drug Discov. 10, 698–712 (2011).
Lue, L. F. et al. Soluble amyloid β peptide concentration as a predictor of synaptic change in Alzheimer's disease. Am. J. Pathol. 155, 853–862 (1999).
Baglioni, S. et al. Prefibrillar amyloid aggregates could be generic toxins in higher organisms. J. Neurosci. 26, 8160–8167 (2006).
Cheon, M. et al. Structural reorganisation and potential toxicity of oligomeric species formed during the assembly of amyloid fibrils. PLoS Comp. Biol. 3, 1727–1738 (2007).
Bolognesi, B. et al. ANS binding reveals common features of cytotoxic amyloid species. ACS Chem. Biol. 5, 735–740 (2010).
Olzscha, H. et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 (2011).
Narayan, P. et al. Single molecule characterization of the interactions between amyloid-β peptides and the membranes of hippocampal cells. J. Am. Chem. Soc. 135, 1491–1498 (2013).
Ciryam, P., Tartaglia, G. G., Morimoto, R. I., Dobson, C. M. & Vendruscolo, M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 5, 781–790 (2013). Provides evidence that links protein supersaturation with ageing and neurodegeneration.
David, D. C. et al. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8, e1000450 (2010).
Gidalevitz, T., Ben-Zvi, A., Ho, K. H., Brignull, H. R. & Morimoto, R. I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 (2006). This work demonstrates how protein homeostasis can be overwhelmed by the additional requests introduced by the presence of folding-defective proteins.
Koga, H., Kaushik, S. & Cuervo, A. M. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res. Rev. 10, 205–215 (2011).
Reis-Rodrigues, P. et al. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell 11, 120–127 (2012).
Cooper-Knock, J. et al. Gene expression profiling in human neurodegenerative disease. Nature Rev. Neurol. 8, 518–530 (2012).
Blalock, E. M. et al. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl Acad. Sci. USA 101, 2173–2178 (2004).
Dobson, M. The story of medicine (Quercus, 2013).
Dobson, C. M. In the footsteps of alchemists. Science 304, 1259–1262 (2004).
Johnson, S. M. et al. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses. Acc. Chem. Res. 38, 911–921 (2005).
Razavi, H. et al. Benzoxazoles as transthyretin amyloid fibril inhibitors: synthesis, evaluation, and mechanism of action. Angew. Chem. Int. Ed. 42, 2758–2761 (2003). Provides initial evidence that stabilizing native states against aggregation offers effective therapeutic interventions.
Bulawa, C. E. et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc. Natl Acad. Sci. USA 109, 9629–9634 (2012).
Uversky, V. N. Intrinsically disordered proteins and novel strategies for drug discovery. Expert Opin. Drug Discov. 7, 475–488 (2012).
Tóth, G. et al. Targeting the intrinsically disordered structural ensemble of α-synuclein by small molecules as a potential therapeutic strategy for Parkinson's disease. PLoS ONE 9, e87133 (2014).
Arosio, P., Vendruscolo, M., Dobson, C. M. & Knowles, T. P. Chemical kinetics for drug discovery to combat protein aggregation diseases. Trends Pharmacol. Sci. 35, 127–135 (2014).
Baigent, C. et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 366, 1267–1278 (2005).
Fersht, A. R. Structure and mechanism in protein science: A guide to enzyme catalysis and protein folding (W. H. Freeman, 1999).
De Strooper, B., Vassar, R. & Golde, T. The secretases: enzymes with therapeutic potential in Alzheimer disease. Nature Rev. Neurol. 6, 99–107 (2010).
Dumoulin, M. & Dobson, C. M. Probing the origins, diagnosis and treatment of amyloid diseases using antibodies. Biochimie 86, 589–600 (2004).
Hard, T. & Lendel, C. Inhibition of amyloid formation. J. Mol. Biol. 421, 441–465 (2012).
Luheshi, L. M. et al. Sequestration of the Aβ peptide prevents toxicity and promotes degradation in vivo. PLoS Biol. 8, e1000334 (2010).
Schenk, D. Amyloid-β immunotherapy for Alzheimer's disease: The end of the beginning. Nature Rev. Neurosci. 3, 824–828 (2002).
Schenk, D., Basi, G. S. & Pangalos, M. N. Treatment strategies targeting amyloid β-protein. Cold Spring Harb. Perspect. Med. 2, a006387 (2012).
Acknowledgements
The authors acknowledge with tremendous gratitude the many graduate students, postdoctoral fellows, collaborators and other colleagues whose discoveries and ideas are reflected in this Review — many of their names are included in the citations to published work. They are also very grateful to the many organizations that have funded their research over many years, including the Wellcome Trust, the Leverhulme Trust, the Alzheimer's Research Trust, Parkinson's UK, the Frances and Augustus Newman Foundation, the European Commission, UK Research Councils and Elan Pharmaceuticals.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Intrinsic
-
In the case of a protein, a property that only depends on its amino acid sequence.
- Protein solubility
-
The concentration of the soluble fraction of a protein in equilibrium with the insoluble fraction. This thermodynamic definition, in the presence of high kinetic barriers between the various soluble and insoluble states of a protein, may, however, need to be extended to include kinetic factors.
- Protein homeostasis
-
The ensemble of cellular processes that regulates the behaviour of proteins in terms of their conformations, interactions, concentrations and localizations.
- Generic
-
In the case of proteins, a property that is common to most of them, as opposed to 'specific' properties. Such generic properties are often associated with the backbone that is common to all polypeptide molecules, whereas specific properties arise from the variations in the chemistry that is mediated by the side chains.
- Chirality
-
The characteristic of a molecular structure that does not have mirror symmetry.
- Young's modulus
-
A measure of the elastic properties of a material, defined as the ratio between stress and strain along a given axis.
- Nucleation
-
In the transition from a fluid phase to a condensed phase, a process that generates species within the fluid phase that are capable of growing into the condensed phase.
- Templating
-
A phenomenon in which structured aggregates promote the conversion of soluble protein species into similar aggregates.
- Seeding
-
A phenomenon in nucleated growth processes by which nuclei of the aggregated phase promote the formation of larger aggregates.
- Primary nucleation
-
A nucleation process that takes place by the spontaneous assembly of monomeric species.
- Secondary nucleation
-
A process by which the formation of new nuclei in the aggregated phase is catalysed by existing aggregates.
- Spreading
-
In the context of neurodegeneration, the spatial propagation of amyloid assemblies from cell to cell by a series of diffusion or transport mechanisms that are coupled with seeding or templating processes.
- Cooperativity
-
A property of a system that results from its collective behaviour, but it is not exhibited by its component parts.
- Molecular chaperones
-
Proteins that assist the protein-folding process, the maintenance of the soluble state of proteins and more generally contribute to generating and preserving protein homeostasis.
- Systemic amyloidoses
-
A group of diseases characterized by the widespread deposition of amyloid aggregates in organs and tissues.
- Protein metastasis
-
The ensemble of molecular processes whereby the functional soluble states of proteins convert into self-propagating aberrant assemblies that initiate a cascade of cytotoxic events.
- Supersaturated
-
A condition in which a soluble substance is concentrated to a level above its critical value but kinetic barriers delay its transition to an insoluble state.
Rights and permissions
About this article
Cite this article
Knowles, T., Vendruscolo, M. & Dobson, C. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol 15, 384–396 (2014). https://doi.org/10.1038/nrm3810
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3810
This article is cited by
-
Maturation-dependent changes in the size, structure and seeding capacity of Aβ42 amyloid fibrils
Communications Biology (2024)
-
Pediococcus pentosaceus LAB6- and Lactiplantibacillus plantarum LAB12-Derived Cell Free Supernatant Inhibited RhoA Activation and Reduced Amyloid-Β In Vitro
Probiotics and Antimicrobial Proteins (2024)
-
Amyloidogenic proteins in the SARS-CoV and SARS-CoV-2 proteomes
Nature Communications (2023)
-
Amyloid formation as a protein phase transition
Nature Reviews Physics (2023)
-
The interface of condensates of the hnRNPA1 low-complexity domain promotes formation of amyloid fibrils
Nature Chemistry (2023)