Abstract
More than 20 years after its discovery, our understanding of target of rapamycin (TOR) signalling continues to grow. Recent global 'omics' studies have revealed physiological roles of mammalian TOR (mTOR) in protein, nucleotide and lipid synthesis. Furthermore, emerging evidence provides new insight into the control of mTOR by other pathways such as Hippo, WNT and Notch signalling. Together, this progress has expanded the list of downstream effectors and upstream regulators of mTOR signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Wullschleger, S., Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Dibble, C. C. & Manning, B. D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nature Cell Biol. 15, 555–564 (2013).
Inoki, K., Li, Y., Zhu, T., Wu, J. & Guan, K. L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature Cell Biol. 4, 648–657 (2002).
Long, X., Lin, Y., Ortiz-Vega, S., Yonezawa, K. & Avruch, J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702–713 (2005).
Ma, X. M. & Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nature Rev. Mol. Cell Biol. 10, 307–318 (2009).
Mamane, Y. Petroulakis, E., LeBacquer, O. & Sonenberg, N. mTOR, translation initiation and cancer. Oncogene 25, 6416–6422 (2006).
Jewell, J. L., Russell, R. C. & Guan, K. L. Amino acid signalling upstream of mTOR. Nature Rev. Mol. Cell Biol. 14, 133–139 (2013).
Mieulet, V. & Lamb, R. F. Tuberous sclerosis complex: linking cancer to metabolism. Trends Mol. Med. 16, 329–335 (2010).
Zinzalla, V., Stracka, D., Oppliger, W. & Hall, M. N. Activation of mTORC2 by association with the ribosome. Cell 144, 757–768 (2011).
Cybulski, N. & Hall, M. N. TOR complex 2: a signaling pathway of its own. Trends Biochem. Sci. 34, 620–627 (2009).
Sparks, C. A. & Guertin, D. A. Targeting mTOR: prospects for mTOR complex 2 inhibitors in cancer therapy. Oncogene 29, 3733–3744 (2010).
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012).
Larsson, O. et al. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc. Natl Acad. Sci. USA 109, 8977–8982 (2012).
Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012).
Hsu, P. P. et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 (2011).
Yu, Y. et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 (2011).
Robitaille, A. M. et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339, 1320–1323 (2013).
Duvel, K. et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010).
Ben-Sahra, I., Howell, J. J., Asara, J. M. & Manning, B. D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339, 1323–1328 (2013).
Chauvin, C. et al. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33, 474–483 (2013).
Raught, B. et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 23, 1761–1769 (2004).
Wang, X. et al. Regulation of elongation factor 2 kinase by p90RSK1 and p70 S6 kinase. EMBO J. 20, 4370–4379 (2001).
Holz, M. K., Ballif, B. A. & Gygi, S. P. & Blenis, J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569–580 (2005).
Mieulet, V. et al. S6 kinase inactivation impairs growth and translational target phosphorylation in muscle cells maintaining proper regulation of protein turnover. Am. J. Physiol. Cell Physiol. 293, C712–722 (2007).
Thoreen, C. C. et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009).
Dowling, R. J. et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328, 1172–1176 (2010).
Oh, W. J. et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 29, 3939–3951 (2010).
Dai, N., Christiansen, J. & Nielsen, F. C. & Avruch, J. mTOR complex 2 phosphorylates IMP1 cotranslationally to promote IGF2 production and the proliferation of mouse embryonic fibroblasts. Genes Dev. 27, 301–312 (2013).
Csibi, A. et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153, 840–854 (2013).
Hagiwara, A. et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell. Metab. 15, 725–738 (2012).
Porstmann, T. et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell. Metab. 8, 224–236 (2008).
Peterson, T. R. et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 (2011).
Sengupta, S., Peterson, T. R., Laplante, M., Oh, S. & Sabatini, D. M. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104 (2010).
Kenerson, H. L., Yeh, M. M. & Yeung, R. S. Tuberous sclerosis complex-1 deficiency attenuates diet-induced hepatic lipid accumulation. PLoS ONE 6, e18075 (2011).
Yecies, J. L. et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell. Metab. 14, 21–32 (2011).
Yuan, M., Pino, E., Wu, L., Kacergis, M. & Soukas, A. A. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J. Biol. Chem. 287, 29579–29588 (2012).
Cornu, M., Albert, V. & Hall, M. N. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 23, 53–62 (2013).
Lamming, D. W. & Sabatini, D. M. A central role for mTOR in lipid homeostasis. Cell. Metab. 18, 465–469 (2013).
Ricoult, S. J. & Manning, B. D. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 14, 242–251 (2013).
Noda, T. & Ohsumi, Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966 (1998).
Blommaart, E. F., Luiken, J. J., Blommaart, P. J., van Woerkom, G. M. & Meijer, A. J. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270, 2320–2326 (1995).
Hosokawa, N. et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009).
Ganley, I. G. et al. ULK1. ATG13. FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009).
Jung, C. H. et al. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009).
Kim, J., Kundu, M., Viollet, B. & Guan, K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biol. 13, 132–141 (2011).
Russell, R. C. et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nature Cell Biol. 15, 741–750 (2013).
Yuan, H. X., Russell, R. C. & Guan, K. L. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 9, 1983–1995 (2013).
Koren, I., Reem, E. & Kimchi, A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 20, 1093–1098 (2010).
Yu, L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010).
Settembre, C. et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012).
Roczniak-Ferguson, A. et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 5, ra42 (2012).
Martina, J. A., Chen, Y., Gucek, M. & Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012).
Martina, J. A. & Puertollano, R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J. Cell Biol. 200, 475–491 (2013).
Pena-Llopis, S. et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30, 3242–3258 (2011).
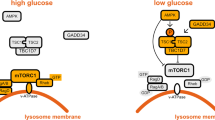
Kim, S. G. et al. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT–RUVBL1/2 complex. Mol. Cell 49, 172–185 (2013).
Zhao, B., Tumaneng, K. & Guan, K. L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nature Cell Biol. 13, 877–883 (2011).
Tumaneng, K. et al. YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nature Cell Biol. 14, 1322–1329 (2012).
Dong, J. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007).
Miller, E. et al. Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem. Biol. 19, 955–962 (2012).
Yu, F. X. et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 (2012).
Wymann, M. P. & Schneiter, R. Lipid signalling in disease. Nature Rev. Mol. Cell Biol. 9, 162–176 (2008).
Niehrs, C. The complex world of WNT receptor signalling. Nature Rev. Mol. Cell Biol. 13, 767–779 (2012).
Inoki, K. et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–968 (2006).
Valvezan, A. J., Huang, J., Lengner, C. J., Pack, M. & Klein, P. S. Oncogenic mutations in adenomatous polyposis coli (Apc) activate mechanistic target of rapamycin complex 1 (mTORC1) in mice and zebrafish. Dis. Model. Mech. 7, 63–71 (2014).
Esen, E. et al. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell. Metab. 17, 745–755 (2013).
Bray, S. J. Notch signalling: a simple pathway becomes complex. Nature Rev. Mol. Cell Biol. 7, 678–689 (2006).
Pajvani, U. B. et al. Inhibition of Notch uncouples Akt activation from hepatic lipid accumulation by decreasing mTorc1 stability. Nature Med. 19, 1054–1060 (2013).
Morita, M. et al. mTORC1 Controls Mitochondrial Activity and Biogenesis through 4E-BP-Dependent Translational Regulation. Cell. Metab. 18, 698–711 (2013).
Betz, C. & Hall, M. N. Where is mTOR and what is it doing there? J. Cell Biol. 203, 563–574 (2013).
Chaveroux, C. et al. Molecular and genetic crosstalks between mTOR and ERRα are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell. Metab. 17, 586–598 (2013).
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008).
Kim, E., Goraksha-Hicks, P., Li, L., Neufeld, T. P. & Guan, K. L. Regulation of TORC1 by Rag GTPases in nutrient response. Nature Cell Biol. 10, 935–945 (2008).
Zoncu, R. et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334, 678–683 (2011).
Sancak, Y. et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010).
Bar-Peled, L., Schweitzer, L. D., Zoncu, R. & Sabatini, D. M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 (2012).
Duran, R. V. et al. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell 47, 349–358 (2012).
Duran, R. V. et al. HIF-independent role of prolyl hydroxylases in the cellular response to amino acids. Oncogene 32, 4549–4556 (2013).
Bar-Peled, L. et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013).
Panchaud, N., Peli-Gulli, M. P. & De Virgilio, C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal 6, ra42 (2013).
Petit, C. S., Roczniak-Ferguson, A. & Ferguson, S. M. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 202, 1107–1122 (2013).
Tsun, Z. Y. et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 (2013).
Benjamin, D., Colombi, M., Moroni, C. & Hall, M. N. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nature Rev. Drug Discov. 10, 868–880 (2011).
Yang, H. et al. mTOR kinase structure, mechanism and regulation. Nature 497, 217–223 (2013).
Kang, S. A. et al. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341, 1236566 (2013).
Acknowledgements
The authors acknowledge support from the Louis-Jeantet Foundation, the Swiss National Science Foundation and the Canton of Basel.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
PowerPoint slides
Rights and permissions
About this article
Cite this article
Shimobayashi, M., Hall, M. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 15, 155–162 (2014). https://doi.org/10.1038/nrm3757
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3757
This article is cited by
-
Altering phosphorylation in cancer through PP2A modifiers
Cancer Cell International (2024)
-
Pannexin 1 targets mitophagy to mediate renal ischemia/reperfusion injury
Communications Biology (2023)
-
Activating Wnt/β-catenin signaling by autophagic degradation of APC contributes to the osteoblast differentiation effect of soy isoflavone on osteoporotic mesenchymal stem cells
Acta Pharmacologica Sinica (2023)
-
Lysosomal control of senescence and inflammation through cholesterol partitioning
Nature Metabolism (2023)
-
Lysosomes mediate the mitochondrial UPR via mTORC1-dependent ATF4 phosphorylation
Cell Discovery (2023)