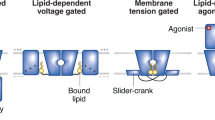
Key Points
-
Ions move across cell membranes through either ion channels or ion pumps. Ions flow passively through ion channels, down electrical and concentration gradients, at speeds that can approach the diffusion limit. By contrast, ion pumps generate those gradients by expending energy (usually in the form of ATP, or gradients of sodium ions or protons) to slowly move ions thermodynamically uphill.
-
So that ions flow only when needed, the pathway through an ion channel can be opened and closed by conformational changes that displace an obstruction called a gate. Although a channel needs only a single gate, a pump needs at least two gates that must open and close strictly alternately to provide access to its binding sites from only one side of the membrane at a time. A pump's two gates should never both be open, lest it behave like an ion channel and allow dissipative ion flow that would in an instant undo the pump's concentrative work.
-
X-ray crystal structures of cation channels and cation pumps perfected by evolution reveal distinct design principles. In ion channels, the narrow region that recognizes and selects ions for conduction is physically separated from the activation gate. But in pumps, these functions are intertwined, as the ion-binding pocket is remodelled with each alternation of access. The need for two gating reactions per transport event accounts for the orders of magnitude slower ion movement through pumps than through channels.
-
High-resolution structures have also been obtained of a member of a family that contains both anion channels and anion pumps. The structures are of a pump, but structural and functional analysis of mutants suggests that only microscopic differences distinguish the two family branches from one another. Moreover, channel members bear vestiges of pump behaviour, implying that the channels evolved from a pump member in which one of the gates failed.
-
There are other examples of ion channels that belong to pump families, and that likely arose from inherited gate dysfunction. Pharmacological interference with gate function can also transform a pump into an ion channel. There are even hybrid molecules that, while pumping their substrate, allow uncoupled, channel-like, electrodiffusive ion leakage.
-
Ion transport proteins can thus span a wide spectrum, from highly evolved ion channels to evolutionarily perfected ion pumps. Somewhere between these extremes, the distinction between channels and pumps becomes blurred. Determining the structures and mechanisms of ion transport proteins throughout the spectrum remains an important challenge.
Abstract
The incessant traffic of ions across cell membranes is controlled by two kinds of border guards: ion channels and ion pumps. Open channels let selected ions diffuse rapidly down electrical and concentration gradients, whereas ion pumps labour tirelessly to maintain the gradients by consuming energy to slowly move ions thermodynamically uphill. Because of the diametrically opposed tasks and the divergent speeds of channels and pumps, they have traditionally been viewed as completely different entities, as alike as chalk and cheese. But new structural and mechanistic information about both of these classes of molecular machines challenges this comfortable separation and forces its re-evaluation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hille, B. Ion Channels of Excitable Membranes (Sinauer, Sunderland, 2001).
Sakmann, B. & Neher, E. Single-Channel Recording (Plenum, New York, 1995).
Läuger, P. A channel mechanism for electrogenic ion pumps. Biochim. Biophys. Acta 552, 143–161 (1979).
Patlak, C. S. Contributions to the theory of active transport. II. The gate-type non-carrier mechanism and generalization concerning tracer glow efficiency, and measurement of energy expenditure. Bull. Math. Biophys. 19, 209–235 (1957).
Vidaver, G. A. Inhibition of parallel flux and augmentation of counter flux shown by transport models not involving a mobile carrier. J. Theor. Biol. 10, 301–306 (1966).
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 27, 969–970 (1966).
MacKinnon, R. Potassium channels. FEBS Lett. 555, 62–65 (2003).
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001). This high-resolution structure revealed the mechanism of the K ion channel selectivity filter. Each of its four K ion sites lies at the centre of a cage of eight carbonyl oxygens that mimic those of the eight water molecules found surrounding the hydrated K ion in the large central cavity. The selectivity filter minimizes the energetic cost of transferring K ions from the aqueous environment to the K ion channel interior.
Morais-Cabral, J. H., Zhou, Y. & MacKinnon, R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature 414, 37–42 (2001).
Post, R. L., Hegyvary, C. & Kume, S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247, 6530–6540 (1972).
Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 (2000). The first X-ray crystal structure of a P-type ATPase ion pump. The two transported Ca ions were found buried side by side in their binding sites, deep within the transmembrane domain and inaccessible from either side.
Toyoshima, C., Nomura, H. & Tsuda, T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature 432, 361–368 (2004).
Toyoshima, C., Norimatsu, Y., Iwasawa, S., Tsuda, T. & Ogawa, H. How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump. Proc. Natl Acad. Sci. USA 104, 19831–19836 (2007).
Toyoshima, C. & Nomura, H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418, 605–611 (2002).
Toyoshima, C. & Mizutani, T. Crystal structure of the calcium pump with a bound ATP analogue. Nature 430, 529–535 (2004).
Møller, J. V., Nissen, P., Sorensen, T. L.-M. & le Marie, M. Transport mechanism of the sarcoplasmic reticulum Ca2+-ATPase pump. Curr. Opin. Struct. Biol. 15, 387–393 (2005).
Olesen, C., Sorensen, T. L., Nielsen, R. C., Møller, J. V. & Nissen, P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science 306, 2251–2255 (2004).
Olesen, C. et al. The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 (2007).
Sørensen, T. L. M., Møller, J. V. & Nissen, P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science 304, 1672–1675 (2004).
Morth, J. P. et al. Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 (2007).
Accardi, A. & Miller, C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature 427, 803–807 (2004). Using electrophysiological and biochemical functional assays on the same preparation of prokaryotic ClC proteins (ClC-ec1) that had been used for X-ray crystallography, this study came to the shocking conclusion that ClC-ec1 is a Cl/H exchange pump, rather than the Cl ion channel initially assumed.
Miller, C. Open-state substructure of single chloride channels from Torpedo electroplax. Philos. Trans. R. Soc. Lond. B Biol. Sci. 299, 401–411 (1982).
Middleton, R. E., Pheasant, D. J. & Miller, C. Homodimeric architecture of a CIC-type chloride ion channel. Nature 383, 337–340 (1996).
Ludewig, U., Pusch, M. & Jentsch, T. J. Two physcially distinct pores in the demeric CIC-0 chloride channel. Nature 383, 340–343 (1996).
Dutzler, R., Campbell, E. B., Cadene, M., Chait, B. T. & MacKinnon, R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294 (2002).
Jentsch, T. J., Poët, M., Fuhrmann, J. C. & Zdebik, A. A. Physiological functions of CLC Cl− channels gleaned from human genetic disease and mouse models. Annu. Rev. Physiol. 67, 779–807 (2005).
Dutzler, R., Campbell, E. B. & MacKinnon, R. Gating the selectivity filter in ClC chloride channels. Science 300, 108–112 (2003). In this structural study, a Glu residue side chain that occupies an anion site in the ion pathway of ClC-ec1 was found to swing away on protonation, unblocking the pathway and allowing a Cl ion to take its place. The residue was identified as a fast gate because mutation of the analogous Glu residue in ClC-0 channels to Gln almost abolished fast gating, leaving the channels mostly open.
Chen, M. F. & Chen, T. Y. Different fast-gate regulation by external Cl− and H+ of the muscle-type ClC chloride channels. J. Gen. Physiol. 118, 23–32 (2001).
Estévez, R., Schroeder, B. C., Accardi, A., Jentsch, T. J. & Pusch, M. Conservation of chloride channel structure revealed by an inhibitor binding site in ClC-1. Neuron 38, 47–59 (2003).
Engh, A. M. & Maduke, M. Cysteine accessibility in ClC-0 supports conservation of the ClC intracellular vestibule. J. Gen. Physiol. 125, 601–617 (2005).
Zifarelli, G. & Pusch, M. ClC chloride channels and transporters: a biophysical and physiological perspective. Rev. Physiol. Biochem. Pharmacol. 158, 23–76 (2007).
Miller, C. ClC chloride channels viewed through a transporter lens. Nature 440, 484–489 (2006).
Jayaram, H., Accardi, A., Wu, F., Williams, C. & Miller, C. Ion permeation through a Cl−-selective channel designed from a ClC Cl−/H+ exchanger. Proc. Natl Acad. Sci. USA 105, 11194–11199 (2008).
Walden, M. et al. Uncoupling and turnover in a Cl−/H+ exchange transporter. J. Gen. Physiol. 129, 317–329 (2007).
Saviane, C., Conti, F. & Pusch, M. The muscle chloride channel ClC-1 has a double-barreled appearance that is differentially affected in dominant and recessive myotonia. J. Gen. Physiol. 113, 457–468 (1999).
Lísal, J. & Maduke, M. The ClC-0 chloride channel is a 'broken' Cl−/H+ antiporter. Nature Struct. Mol. Biol. 15, 805–810 (2008). Using single-channel recordings to show that the gating pattern of ClC-0 channels depends on transmembrane movement of protons, this study firmly established the evolutionary link between ClC pumps and ClC channels and suggested that the channels evolved from a dysfunctional pump.
Gadsby, D. C., Vergani, P. & Csanády, L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature 440, 477–483 (2006).
Muallem, D. & Vergani, P. ATP hydrolysis-driven gating in cystic fibrosis transmembrane conductance regulator. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 247–255 (2009).
Jordan, K., Kota, K. C., Cui, G., Thompson, C. H. & McCarty, N. A. Evolutionary and functional divergence between the cystic fibrosis transmembrane conductance regulator and related ATP-binding cassette transporter. Proc. Natl Acad. Sci. USA. 105, 18865–18870 (2008).
Vergani, P., Lockless, S. W., Nairn, A. C. & Gadsby, D. C. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature 24, 876–880 (2005). This study used CFTR-channel gating kinetics to demonstrate ATP-dependent energetic interaction between residues at positions shown to structurally interact across the interface of prokaryotic nucleotide-binding domain dimers. Evolutionary conservation at these interacting positions in most ABC proteins suggests that nearly all undergo the same ATP-driven cycle of conformational changes established for CFTR.
Nagel, G. et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl Acad. Sci. USA 100, 13940–13945 (2003).
Richard, E. A. & Miller, C. Steady-state coupling of ion-channel conformations to a transmembrane ion gradient. Science 247, 1208–1210 (1990).
Picollo, A. & Pusch, M. Chloride/proton antiporter activity of mammalian ClC proteins ClC-4 and ClC-5. Nature 436, 420–423 (2005).
Riordan, J. R. et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245, 1066–1073 (1989).
Inagaki, N. et al. Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science 270, 1166–1170 (1995).
Hopfner, K. P. et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 (2000). This was the first demonstration that the nucleotide-binding domains of ABC proteins form head-to-tail dimers in the presence of ATP, with one ATP molecule enclosed in each of the two composite catalytic sites formed in the dimer interface. As ADP did not support dimerization, it was proposed that cycles of ATP-induced dimerization and hydrolysis-triggered dissociation provide the basis for function of all ABC proteins.
Sixma, T. K. DNA mismatch repair: MutS structures bound to mismatches. Curr. Opin. Struct. Biol. 11, 47–52 (2001).
Smith, P. C. et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 (2002).
Lu, G., Westbrooks, J. M., Davidson, A. L. & Chen, J. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. Proc. Natl Acad. Sci. USA 102, 17969–17974 (2005).
Dawson, R. J. & Locher, K. P. Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 (2006). This paper reported the first high-resolution structure of an entire ABC transporter that is homologous to clinically relevant human ABC proteins. The structure revealed that the transmembrane helices extend long linker helices into the cytoplasm where they connect to the nucleotide-binding domains via short conserved coupling helices. It also revealed a domain-swapped architecture in which each nucleotide-binding domain receives connections from both N- and C-terminal transmembrane domains.
Dawson, R. J. & Locher, K. P. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 581, 935–938 (2007).
Oldham, M. L., Khare, D., Quiocho, F. A., Davidson, A. L. & Chen, J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450, 515–521 (2007).
Ward, A., Reyes, C. L., Yu, J., Roth, C. B. & Chang, G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl Acad. Sci. USA 104, 19005–19010 (2007).
Linsdell, P. & Hanrahan, J. W. Adenosine triphosphate-dependent asymmetry of anion permeation in the cystic fibrosis transmembrane conductance regulator chloride channel. J. Gen. Physiol. 111, 601–614 (1998).
Kogan, I. et al. CFTR directly mediates nucleotide-regulated glutathione flux. EMBO J. 22, 1981–1989 (2003).
Habermann, E. Palytoxin acts through Na+ K+ ATPase. Toxicon 27, 1171–1187 (1989).
Tosteson, M. T. In Seafood and Freshwater: Toxins Pharmacology, Physiology, and Detection (ed. Botana, L. M.) 549–566 (Marcel Dekker, New York, 2000).
Scheiner-Bobis, G., Meyer zu Heringdorf, D., Christ, M. & Habermann, E. Palytoxin induces K+ efflux from yeast cells expressing the mammalian sodium pump. Mol. Pharmacol. 45, 1132–1136 (1994).
Hirsh, J. K. & Wu, C. H. Palytoxin-induced single-channel currents from the sodium pump synthesized by in vitro expression. Toxicon 35, 169–176 (1997).
Artigas, P. & Gadsby, D. C. Large diameter of palytoxin-induced Na/K pump channels and modulation of palytoxin interaction by Na/K pump ligands. J. Gen. Physiol. 123, 357–376 (2004).
Artigas, P. & Gadsby, D. C. Na+/K+-pump ligands modulate gating of palytoxin-induced ion channels. Proc. Natl Acad. Sci. USA 100, 501–505 (2003). This study provided support for the view of the Na, K pump as a channel controlled by two gates that open strictly alternately. Current recordings showed that palytoxin interferes with this strict coupling, allowing the two gates to sometimes both be open, but that each gate in a palytoxin-bound pump–channel still responds to its physiological ligand, external K ions or cytoplasmic ATP.
Greie, J. C. & Altendorf, K. The K+-translocating KdpFABC complex from Escherichia coli: a P-type ATPase with unique features. J. Bioenerg. Biomembr. 39, 397–402 (2007).
Reyes, N. & Gadsby, D. C. Ion permeation through the Na+, K+-ATPase. Nature 443, 470–474 (2006).
Takeuchi, A., Reyes, N., Artigas, P., Gadsby, D. C. The ion pathway through the opened Na+, K+-ATPase pump. Nature 456, 413–416 (2008).
Gadsby, D. C., Takeuchi, A., Artigas, P. & Reyes, N. Peering into an ATPase ion pump with single-channel recordings. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 229–238 (2009).
Accardi, A. et al. Separate ion pathways in a Cl−/H+ exchanger. J. Gen. Physiol. 126, 563–570 (2005).
Tzingounis, A. V. & Wadiche, J. I. Glutamate transporters: confining runaway excitation by shaping synaptic transmission. Nature Rev. Neurosci. 8, 935–947 (2007).
Torres, G. E. & Amara, S. G. Glutamate and monoamine transporters: new visions of form and function. Curr. Opin. Neurobiol. 17, 304–312 (2007).
Gouaux, E. The molecular logic of sodium-coupled neurotransmitter transporters. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 149–154 (2009).
Fairman, W. A., Vandenberg, R. J., Arriza, J. L., Kavanaugh, M. P. & Amara, S. G. An excitatory amino-acid transporter with properties of a ligand-gated chloride channel. Nature 375, 599–603 (1995).
Larsson, H. P., Picaud, S. A., Werblin, F. S. & Lecar, H. Noise analysis of the glutamate-activated current in photoreceptors. Biophys. J. 70, 733–742 (1996).
Wadiche, J. I. & Kavanaugh, M. P. Macroscopic and microscopic properties of a cloned glutamate transporter/chloride channel. J. Neurosci. 18, 7650–7661 (1998).
Ryan, R. M. & Mindell, J. A. The uncoupled chloride conductance of a bacterial glutamate transporter homolog. Nature Struct. Mol. Biol. 14, 365–371 (2007). This study used reconstituted purified glutamate transporter protein to establish that the uncoupled channel-like Cl current flows through the same protein that carries out stoichiometric Na-dependent transport of glutamate.
Vandenberg, R. J., Huang, S. & Ryan, R. M. Slips, leaks and channels in glutamate transporters. Channels 2, 51–58 (2008).
Ryan, R. M., Mitrovic, A. D. & Vandenberg, R. J. The chloride permeation pathway of a glutamate transporter and its proximity to the glutamate translocation pathway. J. Biol. Chem. 279, 20742–20751 (2004).
Otis, T. S. & Kavanaugh, M. P. Isolation of current components and partial reaction cycles in the glial glutamate transporter EAAT2. J. Neurosci. 20, 2749–2757 (2000).
Post, R. L. & Jolly, P. C. The linkage of sodium, potassium, and ammonium active transport across the human erythrocyte membrane. Biochim. Biophys. Acta 25, 118–128 (1957).
Jørgensen P. L. Isolation of (Na+ plus K+)-ATPase. Methods Enzymol. 32, 277–290 (1974).
Gadsby, D. C., Kimura, J. & Noma, A. Voltage dependence of Na/K pump current in isolated heart cells. Nature 315, 63–65 (1985).
Nakao, M. & Gadsby, D. C. Voltage dependence of Na translocation by the Na/K pump. Nature 323, 628–630 (1986).
Artigas, P. & Gadsby, D. C. Ion channel-like properties of the Na+/K+ pump. Ann. NY Acad. Sci. 976, 31–40 (2002).
Simons, T. J. B. The interaction of ATP-analogues possessing a blocked γ-phosphate group with the sodium pump in human red cells. J. Physiol. 244, 731–739 (1975).
Forbush, B. 3rd. Rapid release of 42K and 86Rb from an occluded state of the Na, K-pump in the presence of ATP or ADP. J. Biol. Chem. 244, 731–739 (1987).
Gadsby D. C. Ion transport: spot the difference. Nature 427, 795–797 (2004).
Lobet, S. & Dutzler, R. Ion-binding properties of the CIC chloride selectivity filter. EMBO J. 25, 24–33 (2006).
Dutzler, R. A structural perspective on CIC channel and transporter function. FEBS Letters 581, 2839–2844 (2007).
Mornon, J. P., Lehn, P. & Callebaut, I. Atomic model of human cystic fibrosis transmembrane conductance regulator: membrane-spanning domains and coupling interfaces. Cell. Mol. Life Sci. 65, 2594–2612 (2008).
Acknowledgements
I thank present and past laboratory members for their contributions to the research and ideas encapsulated here, A. Takeuchi, A. Gulyás-Kovács, P. Vergani, P. Artigas and P. Hoff for help with figures, R. Dutzler and R. MacKinnon for images and the National Institutes of Health for funding via grants HL36783, HL49907 and DK51767.
Author information
Authors and Affiliations
Related links
Glossary
- Membrane potential
-
The difference in electrical potential between one side of a membrane and the other; usually the electrical potential inside a cell measured with respect to that of the extracellular space.
- Activation gate
-
A gate in an ion channel that when opened initiates ion flow through the channel pore.
- Gating reaction
-
A change in the conformation of a transmembrane transport protein that alters access to the substrate translocation pathway.
- Selectivity filter
-
The narrow region of an ion translocation pathway in which the transport protein interacts with the ions to select among them on the basis of their physicochemical characteristics.
Rights and permissions
About this article
Cite this article
Gadsby, D. Ion channels versus ion pumps: the principal difference, in principle. Nat Rev Mol Cell Biol 10, 344–352 (2009). https://doi.org/10.1038/nrm2668
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2668
This article is cited by
-
Bioinspired light-driven chloride pump with helical porphyrin channels
Nature Communications (2024)
-
Light-enhanced osmotic energy generation with an aramid nanofiber membrane
NPG Asia Materials (2023)
-
Nanomechanical action opens endo-lysosomal compartments
Nature Communications (2023)
-
Molecular dynamics study of Cl− permeation through cystic fibrosis transmembrane conductance regulator (CFTR)
Cellular and Molecular Life Sciences (2023)
-
Bionic iontronics based on nano-confined structures
Nano Research (2023)