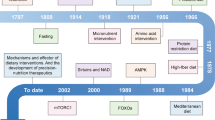
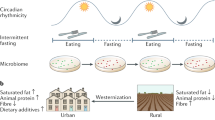
Abstract
Crohn's disease is a chronic relapsing condition that has no certain cure. Both genetic susceptibility and nutrition have key roles, but their level of involvement varies between patients. Interacting gene pathways influence the probability of disease development, but these are affected by stress and various environmental factors, including diet. In addition, the role of the gut microbiome must not be underestimated, as it is substantially altered in patients with Crohn's disease. Although an elemental diet might lead to disease remission, reintroducing real foods and sustainable diets in patients with Crohn's disease is currently difficult, and would benefit from the sensitivity and rapid feedback provided by the field of nutrigenomics. Nutrigenomics utilizes high-throughput genomics technologies to reveal changes in gene and protein expression that are modulated by the patient's nutrition. The most widely used technique thus far is transcriptomics, which permits measurement of changes in the expression of thousands of genes simultaneously in one sample. Given the volume of numbers generated in such studies, data-basing and bioinformatics are essential to ensure the correct application of nutrigenomics at the population level. These methods have been successfully applied to animal models of Crohn's disease, and the time is right to move them to human studies.
Key Points
-
Crohn's disease is an inflammatory condition that develops in genetically predisposed individuals who are exposed to stressors, including certain diets; however, the dietary response cannot currently be predicted with certainty
-
Nutrigenetic, metabonomic and proteomic technologies might enable the identification of patients who will or will not respond to a given diet or medicinal food, thereby increasing the likelihood of efficacy
-
Metabonomics enables the identification of early and sensitive biomarkers that might facilitate validation of novel diets or medicinal foods that could delay disease development or progression
-
Transcriptomics (gene-expression profiling), in combination with advanced bioinformatic methods, might facilitate nonhypothesis-based animal or human trials on the potential effects of certain diets or medicinal foods
-
These methods have proved informative in interpreting the effects of long-chain n-3 fatty acids on Crohn's disease in animal models
-
It is time to apply these methods to develop an improved rationale for dietary interventions in patients with Crohn's disease
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Triggs, C. M. et al. Dietary factors in chronic inflammation: food tolerances and intolerances of a New Zealand Caucasian Crohn's disease population. Mutat. Res. 690, 123–138 (2010).
Ferguson, L. R. et al. Genetic factors in chronic inflammation: single nucleotide polymorphisms in the STAT-JAK pathway, susceptibility to DNA damage and Crohn's disease in a New Zealand population. Mutat. Res. 690, 108–115 (2010).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Rutgeerts, P. J. From aphthous ulcer to full-blown Crohn's disease. Dig. Dis. 29, 211–214 (2011).
Shamir, R. Nutrition and growth in inflammatory bowel disease. J. Pediatr. Gastroenterol. Nutr. 51 (Suppl. 3), S131–S132 (2010).
Petermann, I. et al. Mushroom intolerance: a novel diet-gene interaction in Crohn's disease. Br. J. Nutr. 102, 506–508 (2009).
Wittwer, J. et al. Nutrigenomics in human intervention studies: current status, lessons learned and future perspectives. Mol. Nutr. Food Res. 55, 341–358 (2011).
Afman, L. A. & Müller, M. Nutrigenomics: from molecular nutrition to prevention of disease. J. Am. Diet. Assoc. 106, 569–576 (2006).
Afman, L. A. & Müller, M. Human nutrigenomics of gene regulation by dietary fatty acids. Prog. Lipid Res. 51, 63–70 (2012).
Bakker, G. C. et al. An antiinflammatory dietary mix modulates inflammation and oxidative and metabolic stress in overweight men: a nutrigenomics approach. Am. J. Clin. Nutr. 91, 1044–1059 (2010).
Bouwens, M. et al. Fish-oil supplementation induces antiinflammatory gene expression profiles in human blood mononuclear cells. Am. J. Clin. Nutr. 90, 415–424 (2009).
Roy, N. C., Altermann, E., Park, Z. A. & McNabb, W. C. A comparison of analog and next-generation transcriptomic tools for mammalian studies. Brief. Funct. Genomics 10, 135–150 (2011).
Mesko, B. et al. Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med. Genomics 3, 15 (2010).
Burakoff, R. et al. Blood-based biomarkers can differentiate ulcerative colitis from Crohn's disease and noninflammatory diarrhea. Inflamm. Bowel Dis. 17, 1719–1725 (2011).
Burakoff, R. et al. Differential regulation of peripheral leukocyte genes in patients with active Crohn's disease and Crohn's disease in remission. J. Clin. Gastroenterol. 44, 120–126 (2010).
Lees, C. W., Barrett, J. C., Parkes, M. & Satsangi, J. New IBD genetics: common pathways with other diseases. Gut 60, 1739–1753 (2011).
Hamm, C. M. et al. NOD2 status and human ileal gene expression. Inflamm. Bowel Dis. 16, 1649–1657 (2010).
Lang, M. et al. Gene expression profiles of mucosal fibroblasts from strictured and nonstrictured areas of patients with Crohn's disease. Inflamm. Bowel Dis. 15, 212–223 (2009).
Bogaert, S. et al. Differential mucosal expression of Th17-related genes between the inflamed colon and ileum of patients with inflammatory bowel disease. BMC Immunol. 11, 61 (2010).
Ferguson, L. R., Tatham, A. L., Lin, Z. & Denny, W. A. Epigenetic regulation of gene expression as an anticancer drug target. Curr. Cancer Drug Targets 11, 199–212 (2011).
Konycheva, G. et al. Dietary methyl donor deficiency during pregnancy in rats shapes learning and anxiety in offspring. Nutr. Res. 31, 790–804 (2011).
McKay, J. A. & Mathers, J. C. Diet induced epigenetic changes and their implications for health. Acta Physiol. 202, 103–118 (2011).
Niculescu, M. D. & Lupu, D. S. Nutritional influence on epigenetics and effects on longevity. Curr. Opin. Clin. Nutr. Metab. Care 14, 35–40 (2011).
Zeisel, S. H. Gene response elements, genetic polymorphisms and epigenetics influence the human dietary requirement for choline. IUBMB Life 59, 380–387 (2007).
Ulrich, C. M., Reed, M. C. & Nijhout, H. F. Modeling folate, one-carbon metabolism, and DNA methylation. Nutr. Rev. 66 (Suppl. 1), S27–S30 (2008).
Vujkovic, M. et al. The maternal homocysteine pathway is influenced by riboflavin intake and MTHFR polymorphisms without affecting the risk of orofacial clefts in the offspring. Eur. J. Clin. Nutr. 64, 266–273 (2010).
Carr, D. F., Whiteley, G., Alfirevic, A. & Pirmohamed, M. Investigation of inter-individual variability of the one-carbon folate pathway: a bioinformatic and genetic review. Pharmacogenomics J. 9, 291–305 (2009).
Ma, E. et al. Dietary intake of folate, vitamin B6, and vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer: a case-control study in Brazilian women. BMC Cancer 9, 122 (2009).
Fernández-Miranda, C. et al. Hyperhomocysteinemia and methylenetetrahydrofolate reductase 677C→T and 1298A→C mutations in patients with inflammatory bowel disease. Rev. Esp. Enferm. Dig. 97, 497–504 (2005).
Nakano, E., Taylor, C. J., Chada, L., McGaw, J. & Powers, H. J. Hyperhomocystinemia in children with inflammatory bowel disease. J. Pediatr. Gastroenterol. Nutr. 37, 586–590 (2003).
Mahmud, N. et al. Increased prevalence of methylenetetrahydrofolate reductase C677T variant in patients with inflammatory bowel disease, and its clinical implications. Gut 45, 389–394 (1999).
Oussalah, A., Guéant, J. L. & Peyrin-Biroulet, L. Meta-analysis: hyperhomocysteinaemia in inflammatory bowel diseases. Aliment. Pharmacol. Ther. 34, 1173–1184 (2011).
Stocco, G. et al. Prevalence of methylenetetrahydrofolate reductase polymorphisms in young patients with inflammatory bowel disease. Dig. Dis. Sci. 51, 474–479 (2006).
Zintzaras, E. Genetic variants of homocysteine/folate metabolism pathway and risk of inflammatory bowel disease: a synopsis and meta-analysis of genetic association studies. Biomarkers 15, 69–79 (2010).
Collin, S. M. et al. Association of folate-pathway gene polymorphisms with the risk of prostate cancer: a population-based nested case-control study, systematic review, and meta-analysis. Cancer Epidemiol. Biomarkers Prev. 18, 2528–2539 (2009).
Figueiredo, J. C. et al. Genes involved with folate uptake and distribution and their association with colorectal cancer risk. Cancer Causes Control 21, 597–608 (2010).
Kasperzyk, J. L. et al. Nutrients and genetic variation involved in one-carbon metabolism and Hodgkin lymphoma risk: a population-based case-control study. Am. J. Epidemiol. 174, 816–827 (2011).
Kominsky, D. J. et al. An endogenously anti-inflammatory role for methylation in mucosal inflammation identified through metabolite profiling. J. Immunol. 186, 6505–6514 (2011).
Mowat, C. et al. Guidelines for the management of inflammatory bowel disease in adults. Gut 60, 571–607 (2011).
Nimmo, E. R. et al. Genome-wide methylation profiling in Crohn's disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm. Bowel Dis. http://dx.doi.org/10.1002/ibd.21912.
Ferguson, L. R. RNA silencing: mechanism, biology and responses to environmental stress. Mutat. Res. 714, 93–94 (2011).
Halusková, J. Epigenetic studies in human diseases. Folia Biol. 56, 83–96 (2010).
Richardson, K., Lai, C.-Q., Parnell, L. D., Lee, Y.-C. & Ordovas, J. M. A genome-wide survey for SNPs altering microRNA seed sites identifies functional candidates in GWAS. BMC Genomics 12, 504 (2011).
Jin, G. et al. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis 32, 1655–1659 (2011).
Dimitrov, D. V. The human gutome: nutrigenomics of the host-microbiome interactions. OMICS 15, 419–430 (2011).
Jones, B. V. The human gut mobile metagenome: a metazoan perspective. Gut Microbes 1, 415–431 (2010).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Gentschew, L. & Ferguson, L. R. Role of nutrition and microbiota in susceptibility to inflammatory bowel diseases. Mol. Nutr. Food Res. (in press).
Kang, S. et al. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm. Bowel Dis. 16, 2034–2042 (2010).
Nagalingam, N. A., Kao, J. Y. & Young, V. B. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm. Bowel Dis. 17, 917–926 (2011).
Han, D. Y., Fraser, A. G., Dryland, P. & Ferguson, L. R. Environmental factors in the development of chronic inflammation: a case-control study on risk factors for Crohn's disease within New Zealand. Mutat. Res. 690, 116–122 (2010).
Barnett, M. P. et al. Changes in colon gene expression associated with increased colon inflammation in interleukin-10 gene-deficient mice inoculated with Enterococcus species. BMC Immunol. 11, 39 (2010).
Schaible, T. D., Harris, R. A., Dowd, S. E., Smith, C. W. & Kellermayer, R. Maternal methyl-donor supplementation induces prolonged murine offspring colitis susceptibility in association with mucosal epigenetic and microbiomic changes. Hum. Mol. Genet. 20, 1687–1696 (2011).
van Ommen, B. & Stierum, R. Nutrigenomics: exploiting systems biology in the nutrition and health arena. Curr. Opin. Biotechnol. 13, 517–521 (2002).
de Graaf, A. A. et al. Nutritional systems biology modeling: from molecular mechanisms to physiology. PLoS Comput. Biol. 5, e1000554 (2009).
Funke, B. Laser microdissection of intestinal epithelial cells and downstream analysis. Methods Mol. Biol. 755, 189–196 (2011).
DeBusk, R. The role of nutritional genomics in developing an optimal diet for humans. Nutr. Clin. Pract. 25, 627–633 (2010).
Hurd, P. J. & Nelson, C. J. Advantages of next-generation sequencing versus the microarray in epigenetic research. Brief. Funct. Genomic. Proteomic. 8, 174–183 (2009).
Papanicolaou, A., Stierli, R., Ffrench-Constant, R. H. & Heckel, D. G. Next generation transcriptomes for next generation genomes using est2assembly. BMC Bioinformatics 10, 447 (2009).
Kovatcheva-Datchary, P., Zoetendal, E. G., Venema, K., de Vos, W. M. & Smidt, H. Tools for the tract: understanding the functionality of the gastrointestinal tract. Therap. Adv. Gastroenterol. 2, 9–22 (2009).
Liu, G. E. Applications and case studies of the next-generation sequencing technologies in food, nutrition and agriculture. Recent Pat. Food Nutr. Agric. 1, 75–79 (2009).
Summerer, D. et al. Microarray-based multicycle-enrichment of genomic subsets for targeted next-generation sequencing. Genome Res. 19, 1616–1621 (2009).
Knoch, B. et al. Molecular characterization of the onset and progression of colitis in inoculated interleukin-10 gene-deficient mice: a role for PPARα. PPAR Res. 2010, 621069 (2010).
Dommels, Y. E. et al. Characterization of intestinal inflammation and identification of related gene expression changes in mdr1a−/− mice. Genes Nutr. 2, 209–223 (2007).
Knoch, B. et al. Genome-wide analysis of dietary eicosapentaenoic acid- and oleic acid-induced modulation of colon inflammation in interleukin-10 gene-deficient mice. J. Nutrigenet. Nutrigenomics 2, 9–28 (2009).
Rudkowska, I. et al. Validation of the use of peripheral blood mononuclear cells as surrogate model for skeletal muscle tissue in nutrigenomic studies. OMICS 15, 1–7 (2011).
Anderson, N. L. & Anderson, N. G. Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis 19, 1853–1861 (1998).
Parnell, L. D. & Schueller, C. M. Bioinformatics of the urinary proteome. Methods Mol. Biol. 641, 101–122 (2010).
Kussmann, M., Panchaud, A. & Affolter, M. Proteomics in nutrition: status quo and outlook for biomarkers and bioactives. J. Proteome Res. 9, 4876–4887 (2010).
Bictash, M. et al. Opening up the “Black Box”: metabolic phenotyping and metabolome-wide association studies in epidemiology. J. Clin. Epidemiol. 63, 970–979 (2010).
Chadeau-Hyam, M. et al. Metabolic profiling and the metabolome-wide association study: significance level for biomarker identification. J. Proteome Res. 9, 4620–4627 (2010).
Forsythe, I. J. & Wishart, D. S. Exploring human metabolites using the human metabolome database. Curr. Protoc. Bioinformatic 25, 14.8.1–148.45 (2009).
Nicholson, J. K. & Lindon, J. C. Systems biology: metabonomics. Nature 455, 1054–1056 (2008).
Bjerrum, J. T. et al. Metabonomics in ulcerative colitis: diagnostics, biomarker identification, and insight into the pathophysiology. J. Proteome Res. 9, 954–962 (2010).
Kussmann, M. & Blum, S. OMICS-derived targets for inflammatory gut disorders: opportunities for the development of nutrition related biomarkers. Endocr. Metab. Immune Disord. Drug Targets 7, 271–287 (2007).
Martin, F.-P. et al. Dietary modulation of gut functional ecology studied by fecal metabonomics. J. Proteome Res. 9, 5284–5295 (2010).
McNiven, E. M., German, J. B. & Slupsky, C. M. Analytical metabolomics: nutritional opportunities for personalized health. J. Nutr. Biochem. 22, 995–1002 (2011).
Fay, L. B. & German, J. B. Personalizing foods: is genotype necessary? Curr. Opin. Biotechnol. 19, 121–128 (2008).
Martin, F.-P. et al. Metabolic effects of dark chocolate consumption on energy, gut microbiota, and stress-related metabolism in free-living subjects. J. Proteome Res. 8, 5568–5579 (2009).
Davis, C. D. & Milner, J. A. Nutrigenomics, vitamin D and cancer prevention. J. Nutrigenet. Nutrigenomics 4, 1–11 (2011).
O'Sullivan, A. et al. Biochemical and metabolomic phenotyping in the identification of a vitamin D responsive metabotype for markers of the metabolic syndrome. Mol. Nutr. Food Res. 55, 679–690 (2011).
Cross, H. S., Nittke, T. & Kallay, E. Colonic vitamin D metabolism: Implications for the pathogenesis of inflammatory bowel disease and colorectal cancer. Mol. Cell. Endocrinol. 347, 70–79 (2011).
Chan, E. C. et al. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 8, 352–361 (2009).
Bertini, I. et al. The metabonomic signature of celiac disease. J. Proteome Res. 8, 170–177 (2009).
Lauridsen, M. B. et al. 1H NMR spectroscopy-based interventional metabolic phenotyping: a cohort study of rheumatoid arthritis patients. J. Proteome Res. 9, 4545–4553 (2010).
Lin, H.-M. et al. Metabolomic analysis identifies inflammatory and noninflammatory metabolic effects of genetic modification in a mouse model of Crohn's disease. J. Proteome Res. 9, 1965–1975 (2010).
Lin, H.-M., Helsby, N. A., Rowan, D. D. & Ferguson, L. R. Using metabolomic analysis to understand inflammatory bowel diseases. Inflamm. Bowel Dis. 17, 1021–1029 (2011).
O'Sullivan, A., Gibney, M. J. & Brennan, L. Dietary intake patterns are reflected in metabolomic profiles: potential role in dietary assessment studies. Am. J. Clin. Nutr. 93, 314–321 (2011).
Hearty, A. P. & Gibney, M. J. Comparison of cluster and principal component analysis techniques to derive dietary patterns in Irish adults. Br. J. Nutr. 101, 598–608 (2009).
Heinzmann, S. S. et al. Metabolic profiling strategy for discovery of nutritional biomarkers: proline betaine as a marker of citrus consumption. Am. J. Clin. Nutr. 92, 436–443 (2010).
Dragsted, L. O. Biomarkers of meat intake and the application of nutrigenomics. Meat Sci. 84, 301–307 (2010).
Ferguson, L. R. Meat and cancer. Meat Sci. 84, 308–313 (2010).
Omenn, G. S. Bioinformatics and systems biology of cancers. Prog. Mol. Biol. Transl. Sci. 95, 159–191 (2010).
Yan, Q. Bioinformatics for transporter pharmacogenomics and systems biology: data integration and modeling with UML. Methods Mol. Biol. 637, 23–45 (2010).
Banasik, K. et al. Bioinformatics-driven identification and examination of candidate genes for non-alcoholic fatty liver disease. PLoS ONE 6, e16542 (2011).
Coen, M. et al. Mechanistic aspects and novel biomarkers of responder and non-responder phenotypes in galactosamine-induced hepatitis. J. Proteome Res. 8, 5175–5187 (2009).
Holmes, E., Wilson, I. D. & Nicholson, J. K. Metabolic phenotyping in health and disease. Cell 134, 714–717 (2008).
Huebner, C. et al. Genetic analysis of MDR1 and inflammatory bowel disease reveals protective effect of heterozygous variants for ulcerative colitis. Inflamm. Bowel Dis. 15, 1784–1793 (2009).
Wang, A. H. et al. The effect of IL-10 genetic variation and interleukin 10 serum levels on Crohn's disease susceptibility in a New Zealand population. Hum. Immunol. 72, 431–435 (2011).
Bouwens, M., Grootte Bromhaar, M., Jansen, J., Müller, M. & Afman, L. A. Postprandial dietary lipid-specific effects on human peripheral blood mononuclear cell gene expression profiles. Am. J. Clin. Nutr. 91, 208–217 (2010).
Bouwens, M., Afman, L. A. & Müller, M. Activation of peroxisome proliferator-activated receptor alpha in human peripheral blood mononuclear cells reveals an individual gene expression profile response. BMC Genomics 9, 262 (2008).
Bouwens, M. et al. Fish-oil supplementation induces antiinflammatory gene expression profiles in human blood mononuclear cells. Am. J. Clin. Nutr. 90, 415–424 (2009).
Knoch, B. et al. Genome-wide analysis of dietary eicosapentaenoic acid- and oleic acid-induced modulation of colon inflammation in interleukin-10 gene-deficient mice. J. Nutrigenet. Nutrigenomics 2, 9–28 (2009).
Knoch, B., Nones, K., Barnett, M. P., McNabb, W. C. & Roy, N. C. Diversity of caecal bacteria is altered in interleukin-10 gene-deficient mice before and after colitis onset and when fed polyunsaturated fatty acids. Microbiology 156, 3306–3316 (2010).
Roy, N., Barnett, M., Dommels, Y. & McNabb, W. Nutrigenomics applied to an animal model of inflammatory bowel diseases: transcriptomic analysis of the effects of eicosapentaenoic acid- and arachidonic acid-enriched diets. Mutat. Res. 622, 103–116 (2007).
Knoch, B. et al. Dietary oleic acid as a control fatty acid for polyunsaturated fatty acid intervention studies: a transcriptomics and proteomics investigation using interleukin-10 gene-deficient mice. Biotechnol. J. 5, 1226–1240 (2010).
Cooney, J. M. et al. Proteomic analysis of colon tissue from interleukin-10 gene-deficient mice fed polyunsaturated fatty acids with comparison to transcriptomic analysis. J. Proteome Res. 11, 1065–1077 (2012).
Ferguson, L. R., Smith, B. G. & James, B. J. Combining nutrition, food science and engineering in developing solutions to Inflammatory bowel diseases—omega-3 polyunsaturated fatty acids as an example. Food Funct. 1, 60–72 (2010).
Belluzzi, A. et al. Effect of an enteric-coated fish-oil preparation on relapses in Crohn's disease. N. Engl. J. Med. 334, 1557–1560 (1996).
Feagan, B. G. et al. Omega-3 free fatty acids for the maintenance of remission in Crohn disease: the EPIC Randomized Controlled Trials. JAMA 299, 1690–1697 (2008).
Turner, D., Zlotkin, S. H., Shah, P. S. & Griffiths, A. M. Omega 3 fatty acids (fish oil) for maintenance of remission in Crohn's disease. Cochrane Database of Systematic Reviews, Issue 1. Art. No.: CD006320. http://dx.doi.org/10.1002/14651858.CD006320.pub3.
Williams, C. M. et al. The challenges for molecular nutrition research 1: linking genotype to healthy nutrition. Genes Nutr. 3, 41–49 (2008).
van Ommen, B. et al. The challenges for molecular nutrition research 2: quantification of the nutritional phenotype. Genes Nutr. 3, 51–59 (2008).
Daniel, H., Drevon, C. A., Klein, U. I., Kleemann, R. & van Ommen, B. The challenges for molecular nutrition research 3: comparative nutrigenomics research as a basis for entering the systems level. Genes Nutr. 3, 101–106 (2008).
van Ommen, B., Cavallieri, D., Roche, H. M., Klein, U. I. & Daniel, H. The challenges for molecular nutrition research 4: the “nutritional systems biology level”. Genes Nutr. 3, 107–113 (2008).
van Ommen, B. et al. Challenges of molecular nutrition research 6: the nutritional phenotype database to store, share and evaluate nutritional systems biology studies. Genes Nutr. 5, 189–203 (2010).
Acknowledgements
I wish to thank Virginia Parslow, for editorial help, and Valerie Gray, for the illustrations. Nutrigenomics New Zealand (www.nutrigenomics.org.nz) is a collaboration among The University of Auckland, Plant and Food Research Ltd and AgResearch Ltd. It is funded by the New Zealand Ministry of Science and Innovation.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Ferguson, L. Potential value of nutrigenomics in Crohn's disease. Nat Rev Gastroenterol Hepatol 9, 260–270 (2012). https://doi.org/10.1038/nrgastro.2012.41
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2012.41