Key Points
-
Whole-exome and whole-genome sequencing have provided a comprehensive and high-resolution view of somatic genomic alterations in liver cancer
-
Global epigenetic analyses have further identified both unique and complementary molecular alterations in liver cancer
-
Somatic mutational signatures of the liver cancer genome are complex and tend to be associated with epidemiological backgrounds
-
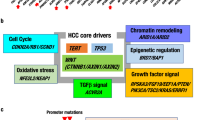
Integration of genetic and epigenetic alteration profiles has elucidated core oncogenic pathways, potential therapeutic targets and new molecular classifications in liver cancer
Abstract
Liver cancer is the third leading cause of cancer-related death worldwide. Advances in sequencing technologies have enabled the examination of liver cancer genomes at high resolution; somatic mutations, structural alterations, HBV integration, RNA editing and retrotransposon changes have been comprehensively identified. Furthermore, integrated analyses of trans-omics data (genome, transcriptome and methylome data) have identified multiple critical genes and pathways implicated in hepatocarcinogenesis. These analyses have uncovered potential therapeutic targets, including growth factor signalling, WNT signalling, the NFE2L2-mediated oxidative pathway and chromatin modifying factors, and paved the way for new molecular classifications for clinical application. The aetiological factors associated with liver cancer are well understood; however, their effects on the accumulation of somatic changes and the influence of ethnic variation in risk factors still remain unknown. The international collaborations of cancer genome sequencing projects are expected to contribute to an improved understanding of risk evaluation, diagnosis and therapy for this cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jemal, A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011).
Forner, A., Llovet, J. M. & Bruix, J. Hepatocellular carcinoma. Lancet 379, 1245–1255 (2012).
El-Serag, H. B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142, 1264–1273 (2012).
Yu, J., Shen, J., Sun, T. T., Zhang, X. & Wong, N. Obesity, insulin resistance, NASH and hepatocellular carcinoma. Semin. Cancer Biol. 23, 483–491 (2013).
Shaib, Y. & El-Serag, H. B. The epidemiology of cholangiocarcinoma. Semin. Liver Dis. 24, 115–125 (2004).
Palmer, W. C. & Patel, T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J. Hepatol. 57, 69–76 (2012).
Takeo, S. et al. Examination of oncogene amplification by genomic DNA microarray in hepatocellular carcinomas: comparison with comparative genomic hybridization analysis. Cancer Genet. Cytogenet. 130, 127–132 (2001).
Yasui, K. et al. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology 35, 1476–1484 (2002).
Okamoto, H., Yasui, K., Zhao, C., Arii, S. & Inazawa, J. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology 38, 1242–1249 (2003).
Patil, M. A. et al. Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis 26, 2050–2057 (2005).
Midorikawa, Y. et al. Molecular karyotyping of human hepatocellular carcinoma using single-nucleotide polymorphism arrays. Oncogene 25, 5581–5590 (2006).
Poon, T. C. et al. A tumor progression model for hepatocellular carcinoma: bioinformatic analysis of genomic data. Gastroenterology 131, 1262–1270 (2006).
Katoh, H. et al. Genetically distinct and clinically relevant classification of hepatocellular carcinoma: putative therapeutic targets. Gastroenterology 133, 1475–1486 (2007).
Ma, N. F. et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology 47, 503–510 (2008).
Schlaeger, C. et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology 47, 511–520 (2008).
Lee, S. A. et al. Integration of genomic analysis and in vivo transfection to identify sprouty 2 as a candidate tumor suppressor in liver cancer. Hepatology 47, 1200–1210 (2008).
Chochi, Y. et al. A copy number gain of the 6p arm is linked with advanced hepatocellular carcinoma: an array-based comparative genomic hybridization study. J. Pathol. 217, 677–684 (2009).
Guo, X. et al. A meta-analysis of array-CGH studies implicates antiviral immunity pathways in the development of hepatocellular carcinoma. PLoS ONE 6, e28404 (2011).
Wang, K. et al. Genomic landscape of copy number aberrations enables the identification of oncogenic drivers in hepatocellular carcinoma. Hepatology 58, 706–717 (2013).
Roessler, S. et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 142, 957–966 (2012).
Zender, L. et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 (2008).
Sawey, E. T. et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell 19, 347–358 (2011).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 39, 1522–1527 (2007).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009).
Li, M. et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 (2011).
Guichard, C. et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 44, 694–698 (2012).
Huang, J. et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat. Genet. 44, 1117–1121 (2012).
Cleary, S. P. et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology http://dx.doi.org/10.1002/hep.26540.
Ong, C. K. et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat. Genet. 44, 690–693 (2012).
Totoki, Y. et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 43, 464–469 (2011).
Fujimoto, A. et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 44, 760–764 (2012).
Kan, Z. et al. Whole genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 23, 1422–1433 (2013).
Neuveut, C., Wei, Y. & Buendia, M. A. Mechanisms of HBV-related hepatocarcinogenesis. J. Hepatol. 52, 594–604 (2010).
Jiang, Z. et al. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 22, 593–601 (2012).
Sung, W. K. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 44, 765–769 (2012).
Treangen, T. J. & Salzberg, S. L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13, 36–46 (2011).
Shukla, R. et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153, 101–111 (2013).
Stratton, M. R., Campbell, P. J. & Futreal, P. A. The cancer genome. Nature 458, 719–724 (2009).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Holczbauer, A. et al. Modeling pathogenesis of primary liver cancer in lineage-specific mouse cell types. Gastroenterology 145, 221–231 (2013).
Herceg, Z. & Paliwal, A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat. Res. 727, 55–61 (2011).
Eden, A., Gaudet, F., Waghmare, A. & Jaenisch, R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300, 455 (2003).
Nagae, G. et al. Tissue-specific demethylation in CpG-poor promoters during cellular differentiation. Hum. Mol. Genet. 20, 2710–2721 (2011).
Toyota, M. et al. CpG island methylator phenotype in colorectal cancer. Proc. Natl Acad. Sci. USA 96, 8681–8686 (1999).
Tao, M. H. & Freudenheim, J. L. DNA methylation in endometrial cancer. Epigenetics 5, 491–498 (2010).
Turcan, S. et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012).
Sato, Y. et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 45, 860–867 (2013).
Herath, N. I., Leggett, B. A. & MacDonald, G. A. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J. Gastroenterol. Hepatol. 21, 15–21 (2006).
Zhang, C. et al. CpG island methylator phenotype association with elevated serum alpha-fetoprotein level in hepatocellular carcinoma. Clin. Cancer Res. 13, 944–952 (2007).
Pastor, W. A., Aravind, L. & Rao, A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 (2013).
Kudo, Y. et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 103, 670–676 (2012).
Wang, P. et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 32, 3091–3100 (2013).
Noushmehr, H. et al. Identification of a CpG Island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 (2010).
Deng, Y. B. et al. Identification of genes preferentially methylated in hepatitis C virus-related hepatocellular carcinoma. Cancer Sci. 101, 1501–1510 (2010).
Bibikova, M. et al. Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics 1, 177–200 (2009).
Clark, C. et al. A comparison of the whole genome approach of MeDIP-seq to the targeted approach of the Infinium HumanMethylation450 BeadChip® for methylome profiling. PLoS ONE 7, e50233 (2012).
Ogino, S. et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod. Pathol. 26, 465–484 (2013).
Shen, J. et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology 55, 1799–1808 (2012).
Shen, J. et al. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics 8, 34–43 (2013).
Tao, R. et al. Methylation profile of single hepatocytes derived from hepatitis B virus-related hepatocellular carcinoma. PLoS ONE 6, e19862 (2011).
Ushijima, T. Epigenetic field for cancerization. J. Biochem. Mol. Biol. 40, 142–150 (2007).
Nishida, N. et al. Extensive methylation is associated with beta-catenin mutations in hepatocellular carcinoma: evidence for two distinct pathways of human hepatocarcinogenesis. Cancer Res. 67, 4586–4594 (2007).
Zang, J. J. et al. P16 gene hypermethylation and hepatocellular carcinoma: a systematic review and meta-analysis. World J. Gastroenterol. 17, 3043–3048 (2011).
Wong, I. H. et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 59, 71–73 (1999).
Narimatsu, T. et al. p16 promoter hypermethylation in human hepatocellular carcinoma with or without hepatitis virus infection. Intervirology 47, 26–31 (2004).
Zhang, Y. J. et al. High frequency of promoter hypermethylation of the RASSF1A and p16 genes and its relationship to aflatoxin B1-DNA adducts level in human hepatocellular carcinoma. Mol. Carcinogenesis 35, 85–92 (2002).
Zhong, S. et al. Silencing of GSTP1 gene by CpG island DNA hypermethylation in HBV-associated hepatocellular carcinomas. Clin. Cancer Res. 8, 1087–1092 (2002).
Yang, B., Guo, M., Herman, J. G. & Clark, D. P. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am. J. Pathol. 163, 1101–1107 (2003).
Zhang, Y. J. et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relationship to aflatoxin B1-DNA adducts and p53 mutations in hepatocellular carcinoma. Int. J. Cancer 103, 440–444 (2003).
Boyault, S. et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 45, 42–52 (2007).
Hoshida, Y. et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 69, 7385–7392 (2009).
Midorikawa, Y. et al. Allelic imbalances and homozygous deletion on 8p23.2 for stepwise progression of hepatocarcinogenesis. Hepatology 49, 513–522 (2009).
Vetter, D. et al. Enhanced hepatocarcinogenesis in mouse models and human hepatocellular carcinoma by coordinate KLF6 depletion and increased messenger RNA splicing. Hepatology 56, 1361–1370 (2012).
Berasain, C. et al. Impairment of pre-mRNA splicing in liver disease: mechanisms and consequences. World J. Gastroenterol. 16, 3091–3102 (2010).
Lu, X. et al. Aberrant splicing of Hugl-1 is associated with hepatocellular carcinoma progression. Clin. Cancer Res. 15, 3287–3296 (2009).
Tsedensodnom, O. et al. Identification of T-cell factor-4 isoforms that contribute to the malignant phenotype of hepatocellular carcinoma cells. Exp. Cell Res. 317, 920–931 (2011).
Castillo, J. et al. Amphiregulin induces the alternative splicing of p73 into its oncogenic isoform DeltaEx2p73 in human hepatocellular tumors. Gastroenterology 137, 1805–1815 (2009).
Li, Y., Chen, L., Chan, T. H. & Guan, X. Y. Hepatocellular carcinoma: transcriptome diversity regulated by RNA editing. Int. J. Biochem. Cell Biol. 45, 1843–1848 (2013).
Chen, L. et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19, 209–216 (2013).
Hussain, S. P. et al. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene 26, 2166–2176 (2007).
Shiloh, Y. & Ziv, Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210 (2013).
el-Deiry, W. S. et al. WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 (1993).
Liew, C. T. et al. High frequency of p16INK4A gene alterations in hepatocellular carcinoma. Oncogene 18, 789–795 (1999).
Zhang, C. et al. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. Int. J. Cancer 123, 998–1004 (2008).
Mayhew, C. N. et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology 133, 976–984 (2007).
Kim, N. W. et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994).
Huang, F. W. et al. Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013).
Horn, S. et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013).
Killela, P. J. et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl Acad. Sci. USA 110, 6021–6026 (2013).
Nault, J. C. et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 4, 2218 (2013).
Polakis, P. Wnt signaling and cancer. Genes Dev. 14, 1837–1851 (2000).
de La Coste, A. et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl Acad. Sci. USA 95, 8847–8851 (1998).
Satoh, S. et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat. Genet. 24, 245–250 (2000).
Oda, H., Imai, Y., Nakatsuru, Y., Hata, J. & Ishikawa, T. Somatic mutations of the APC gene in sporadic hepatoblastomas. Cancer Res. 56, 3320–3323 (1996).
Taniguchi, K. et al. Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene 21, 4863–4871 (2002).
Takagi, H. et al. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J. Gastroenterol. 43, 378–389 (2008).
Tsao, C. M. et al. SOX1 functions as a tumor suppressor by antagonizing the WNT/β-catenin signaling pathway in hepatocellular carcinoma. Hepatology 56, 2277–2287 (2012).
Lévy, L., Renard, C. A., Wei, Y. & Buendia, M. A. Genetic alterations and oncogenic pathways in hepatocellular carcinoma. Ann. NY Acad. Sci. 963, 21–36 (2002).
He, T. C. et al. Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 (1998).
Tetsu, O. & McCormick, F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 (1999).
Euskirchen, G., Auerbach, R. K. & Snyder, M. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 287, 30897–30905 (2012).
Wilson, B. G. & Roberts, C. W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 (2011).
Shain, A. H. & Pollack, J. R. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE 8, e55119 (2013).
Chi, P., Allis, C. D. & Wang, G. G. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010).
Milne, T. A. et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117 (2002).
Ruthenburg, A. J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011).
Yoshikawa, H. et al. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat. Genet. 28, 29–35 (2001).
Challen, C., Guo, K., Collier, J. D., Cavanagh, D., Bassendine, M. F. Infrequent point mutations in codons 12 and 61 of ras oncogenes in human hepatocellular carcinomas. J. Hepatol. 14, 342–346 (1992).
Tanaka, Y. et al. Absence of PIK3CA hotspot mutations in hepatocellular carcinoma in Japanese patients. Oncogene 25, 2950–2952 (2006).
Andersen, J. B. et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 142, 1021–1031 (2012).
Wu, K. et al. Hepatic transforming growth factor beta gives rise to tumor-initiating cells and promotes liver cancer development. Hepatology 56, 2255–2267 (2012).
Nussbaum, T. et al. Autocrine insulin-like growth factor-II stimulation of tumor cell migration is a progression step in human hepatocarcinogenesis. Hepatology 48, 146–156 (2008).
Yoshiji, H. et al. Vascular endothelial growth factor tightly regulates in vivo development of murine hepatocellular carcinoma cells. Hepatology 28, 1489–1496 (1998).
Taguchi, K., Motohashi, H. & Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 16, 123–140 (2011).
Shibata, T. et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl Acad. Sci. USA 105, 13568–13573 (2008).
Mitsuishi, Y. et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22, 66–79 (2012).
Tschaharganeh, D. F. et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology 144, 1530–1542 (2013).
Villanueva, A. et al. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology 143, 1660–1669 (2012).
Viatour, P. et al. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J. Exp. Med. 208, 1963–1976 (2011).
Qi, R. et al. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 63, 8323–8329 (2003).
Radtke, F. & Raj, K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer 3, 756–767 (2003).
Schirmacher, P. & Calvisi, D. F. Molecular diagnostic algorithms in hepatocellular carcinoma: dead-end street or light at the end of the tunnel? Gastroenterology 145, 49–53 (2013).
Boyault, S. et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 45, 42–52 (2007).
Hoshida, Y. et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 69, 7385–7392 (2009).
Chiang, D. Y. et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 68, 6779–6788 (2008).
Imamura, H. et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J. Hepatol. 38, 200–207 (2003).
Hoshida, Y. et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N. Engl. J. Med. 359, 1995–2004 (2008).
Nault, J. C. et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology 145, 176–187 (2013).
Acknowledgements
We apologize for the many excellent works which are not acknowledged in the reference list owing to the limited space. The work of the authors is supported partially by Grants-in-Aid from the Ministry of Health, Labour and Welfare for the 3rd-term Comprehensive 10-year Strategy for Cancer Control, National Cancer Center Research and Development Fund (23-A-8) (T. Shibata), and JSPS KAKENHI Grant Number 24221011 (A. Aburatani).
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to all aspects of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Shibata, T., Aburatani, H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol 11, 340–349 (2014). https://doi.org/10.1038/nrgastro.2014.6
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2014.6
This article is cited by
-
An integrated investigation of sulfotransferases (SULTs) in hepatocellular carcinoma and identification of the role of SULT2A1 on stemness
Apoptosis (2024)
-
An overview of mouse models of hepatocellular carcinoma
Infectious Agents and Cancer (2023)
-
Synthetic miR-26a mimics delivered by tumor exosomes repress hepatocellular carcinoma through downregulating lymphoid enhancer factor 1
Hepatology International (2023)
-
Overview and clinical significance of multiple mutations in individual genes in hepatocellular carcinoma
BMC Cancer (2022)
-
Fabrication of electrospun gum Arabic–polyvinyl alcohol blend nanofibers for improved viability of the probiotic
Journal of Food Science and Technology (2022)