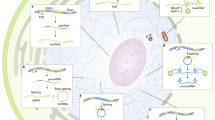
Key Points
-
Nonsense-mediated mRNA decay (NMD) largely provides a means to ensure the quality of gene expression by eliminating mRNAs that prematurely terminate translation either because of genomic mutations or errors in cellular processes.
-
NMD is also crucial to regulating proper expression levels for certain genes and for maintaining genome stability.
-
Up-frameshift (UPF) and phosphatidyl-inositol 3-kinase-related protein kinase (PIKK) SMG proteins are NMD factors, and incompletely defined combinations of these proteins demonstrate unexpected roles in pathways that have no apparent connection to NMD.
-
UPF1, which is an RNA-dependent ATPase and RNA helicase, functions in specialized pathways of mRNA decay, including Staufen 1 (STAU1)-mediated mRNA decay and the degradation of replication-dependent histone mRNAs at the end of the S phase of the cell cycle.
-
Apart from its role in histone mRNA decay, UPF1 influences DNA synthesis and cell-cycle progression by binding to chromatin in a way that is upregulated by the PIKKs ataxia-telangiectasia mutated and Rad 3-related (ATR), SMG1, and ataxia-telangiectasia mutated (ATM). Furthermore, UPF1 seems to be a component of DNA synthesis and repair pathways because it co-immunopurifies with DNA polymerase δ.
-
SMG1 has a role in stress-response pathways and DNA repair triggered by, for example, ionizing radiation, ultraviolet-B light or hyperoxia.
-
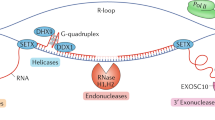
SMG and UPF factors function in the maintenance of telomeres at the ends of chromosomes by inhibiting the binding of telomeric repeat-containing RNA (TERRA) to telomeres.
-
In summary, SMG and UPF proteins can be viewed as components of overarching RNA and DNA surveillance processes and as multitasking players in the intricate network of signal transduction pathways that respond to genetic and acquired errors in nucleic-acid metabolism.
Abstract
Nonsense-mediated mRNA decay (NMD) largely functions to ensure the quality of gene expression. However, NMD is also crucial to regulating appropriate expression levels for certain genes and for maintaining genome stability. Furthermore, just as NMD serves cells in multiple ways, so do its constituent proteins. Recent studies have clarified that UPF and SMG proteins, which were originally discovered to function in NMD, also have roles in other pathways, including specialized pathways of mRNA decay, DNA synthesis and cell-cycle progression, and the maintenance of telomeres. These findings suggest a delicate balance of metabolic events — some not obviously related to NMD — that can be influenced by the cellular abundance, location and activity of NMD factors and their binding partners.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Behm-Ansmant, I. et al. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 581, 2845–2853 (2007).
Chang, Y. F., Imam, J. S. & Wilkinson, M. F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51–74 (2007).
Isken, O. & Maquat, L. E. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21, 1833–1856 (2007). A comprehensive review of translation-dependent mRNA surveillance mechanisms, with a focus on NMD.
Holbrook, J. A., Neu-Yilik, G., Hentze, M. W. & Kulozik, A. E. Nonsense-mediated decay approaches the clinic. Nature Genet. 36, 801–808 (2004).
Amrani, N., Sachs, M. S. & Jacobson, A. Early nonsense: mRNA decay solves a translational problem. Nature Rev. Mol. Cell Biol. 7, 415–425 (2006). A comprehensive overview of NMD in S. cerevisiae
Meaux, S., van Hoof, A. & Baker, K. E. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol. Cell 29, 134–140 (2008).
Chiu, S. Y., Lejeune, F., Ranganathan, A. C. & Maquat, L. E. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 18, 745–754 (2004).
Lejeune, F., Ranganathan, A. C. & Maquat, L. E. eIF4G is required for the pioneer round of translation in mammalian cells. Nature Struct. Mol. Biol. 11, 992–1000 (2004).
Sato, H., Hosoda, N. & Maquat, L. E. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol. Cell 29, 255–262 (2008).
Matsuda, D., Hosoda, N., Kim, Y. K. & Maquat, L. E. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nature Struct. Mol. Biol. 14, 974–979 (2007).
Hosoda, N., Lejeune, F. & Maquat, L. E. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 26, 3085–3097 (2006).
Woeller, C. F., Gaspari, M., Isken, O. & Maquat, L. E. NMD resulting from encephalomyocarditis virus IRES-directed translation initiation seems to be restricted to CBP80/20-bound mRNA. EMBO Rep. 9, 446–451 (2008).
Ishigaki, Y., Li, X., Serin, G. & Maquat, L. E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617 (2001).
Lejeune, F., Ishigaki, Y., Li, X. & Maquat, L. E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 21, 3536–3545 (2002).
Hosoda, N., Kim, Y. K., Lejeune, F. & Maquat, L. E. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nature Struct. Mol. Biol. 12, 893–901 (2005).
Isken, O. et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133, 314–327 (2008).
Fortes, P. et al. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6, 191–196 (2000).
Das, B., Guo, Z., Russo, P., Chartrand, P. & Sherman, F. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 20, 2827–2838 (2000).
Baron-Benhamou, J., Fortes, P., Inada, T., Preiss, T. & Hentze, M. W. The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast. RNA 9, 654–662 (2003).
Gao, Q., Das, B., Sherman, F. & Maquat, L. E. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc. Natl Acad. Sci. USA 102, 4258–4263 (2005).
Park, N. I. & Muench, D. G. Biochemical and cellular characterization of the plant ortholog of PYM, a protein that interacts with the exon junction complex core proteins Mago and Y14. Planta 225, 625–639 (2007).
Ballut, L. et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nature Struct. Mol. Biol. 12, 861–869 (2005).
Tange, T. O., Shibuya, T., Jurica, M. S. & Moore, M. J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11, 1869–1883 (2005).
Gatfield, D. & Izaurralde, E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159, 579–588 (2002).
Longman, D., Johnstone, I. L. & Caceres, J. F. The REF/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 9, 881–891 (2003).
Kawano, T., Kataoka, N., Dreyfuss, G. & Sakamoto, H. Ce-Y14 and MAG-1, components of the exon–exon junction complex, are required for embryogenesis and germline sexual switching in Caenorhabditis elegans. Mech. Dev. 121, 27–35 (2004).
Gatfield, D., Unterholzner, L., Ciccarelli, F. D., Bork, P. & Izaurralde, E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22, 3960–3970 (2003).
Longman, D., Plasterk, R. H., Johnstone, I. L. & Caceres, J. F. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 21, 1075–1085 (2007).
Serin, G., Gersappe, A., Black, J. D., Aronoff, R. & Maquat, L. E. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol. Cell. Biol. 21, 209–223 (2001).
Gehring, N. H., Neu-Yilik, G., Schell, T., Hentze, M. W. & Kulozik, A. E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11, 939–949 (2003).
Kunz, J. B., Neu-Yilik, G., Hentze, M. W., Kulozik, A. E. & Gehring, N. H. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12, 1015–1022 (2006).
Tarpey, P. S. et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nature Genet. 39, 1127–1133 (2007).
Lykke-Andersen, J., Shu, M. D. & Steitz, J. A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131 (2000).
Mendell, J. T., Medghalchi, S. M., Lake, R. G., Noensie, E. N. & Dietz, H. C. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 20, 8944–8957 (2000).
Mendell, J. T., ap Rhys, C. M. & Dietz, H. C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298, 419–422 (2002).
Gehring, N. H. et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell 20, 65–75 (2005).
Chan, W. K. et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 26, 1820–1830 (2007).
Saltzman, A. et al. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 28, 4320–4330 (2008).
Kashima, I. et al. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20, 355–367 (2006).
Ivanov, P. V., Gehring, N. H., Kunz, J. B., Hentze, M. W. & Kulozik, A. E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27, 736–747 (2008).
Amrani, N. et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432, 112–118 (2004).
Wittmann, J., Hol, E. M. & Jack, H. M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 26, 1272–1287 (2006).
Ohnishi, T. et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol. Cell 12, 1187–1200 (2003).
Chamieh, H., Ballut, L., Bonneau, F. & Le Hir, H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nature Struct. Mol. Biol. 15, 85–93 (2008).
Yamashita, A., Ohnishi, T., Kashima, I., Taya, Y. & Ohno, S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15, 2215–2228 (2001).
Grimson, A., O'Connor, S., Newman, C. L. & Anderson, P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol. 24, 7483–7490 (2004).
Unterholzner, L. & Izaurralde, E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell 16, 587–596 (2004).
Glavan, F., Behm-Ansmant, I., Izaurralde, E. & Conti, E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 25, 5117–5125 (2006).
Brumbaugh, K. M. et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell 14, 585–598 (2004). This paper implicates SMG1, the newest member of the family of protein serine-threonine kinases, in stress-induced signalling pathways.
Page, M. F., Carr, B., Anders, K. R., Grimson, A. & Anderson, P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 19, 5943–5951 (1999).
Johns, L., Grimson, A., Kuchma, S. L., Newman, C. L. & Anderson, P. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol. Cell. Biol. 27, 5630–5638 (2007).
Chen, Z., Smith, K. R., Batterham, P. & Robin, C. Smg1 nonsense mutations do not abolish nonsense-mediated mRNA decay in Drosophila melanogaster. Genetics 171, 403–406 (2005).
Metzstein, M. M. & Krasnow, M. A. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2, e180 (2006).
Wang, W., Cajigas, I. J., Peltz, S. W., Wilkinson, M. F. & Gonzalez, C. I. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol. Cell. Biol. 26, 3390–3400 (2006).
Luke, B. et al. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res. 35, 7688–7697 (2007).
Lynch, M., Hong, X. & Scofield, D. S. in Nonsense-Mediated mRNA Decay (ed. L. E. Maquat) 197–211 (Landes Bioscience, Georgetown, 2006). A beautifully written treatise on the association of NMD, the EJC and the location of introns within genes on the basis of phylogenetic analyses.
Lelivelt, M. J. & Culbertson, M. R. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 19, 6710–6719 (1999).
He, F. et al. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12, 1439–1452 (2003).
Mendell, J. T., Sharifi, N. A., Meyers, J. L., Martinez-Murillo, F. & Dietz, H. C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature Genet. 36, 1073–1078 (2004).
Rehwinkel, J., Letunic, I., Raes, J., Bork, P. & Izaurralde, E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11, 1530–1544 (2005).
Rodriguez-Gabriel, M. A., Watt, S., Bahler, J. & Russell, P. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol. Cell. Biol. 26, 6347–6356 (2006).
Johansson, M. J., He, F., Spatrick, P., Li, C. & Jacobson, A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc. Natl Acad. Sci. USA 104, 20872–20877 (2007). This paper and reference 63 use microarray analyses to identify direct targets of NMD in S. cerevisiae.
Guan, Q. et al. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2, e203 (2006).
Moriarty, P. M., Reddy, C. C. & Maquat, L. E. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 18, 2932–2939 (1998).
Wen, W., Weiss, S. L. & Sunde, R. A. UGA codon position affects the efficiency of selenocysteine incorporation into glutathione peroxidase-1. J. Biol. Chem. 273, 28533–28541 (1998).
Mitrovich, Q. M. & Anderson, P. mRNA surveillance of expressed pseudogenes in C. elegans. Curr. Biol. 15, 963–967 (2005).
Ni, J. Z. et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 21, 708–718 (2007). In parallel with reference 76, this paper identifies RNA-binding transcripts containing ultraconserved sequences that are subject to alternative splicing-activated NMD.
Weischenfeldt, J. et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 22, 1381–1396 (2008).
Lewis, B. P., Green, R. E. & Brenner, S. E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA 100, 189–192 (2003).
Sureau, A., Gattoni, R., Dooghe, Y., Stevenin, J. & Soret, J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 20, 1785–1796 (2001).
Wollerton, M. C., Gooding, C., Wagner, E. J., Garcia-Blanco, M. A. & Smith, C. W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100 (2004).
Jumaa, H. & Nielsen, P. J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 16, 5077–5085 (1997).
Lejeune, F., Cavaloc, Y. & Stevenin, J. Alternative splicing of intron 3 of the serine/arginine-rich protein 9G8 gene. Identification of flanking exonic splicing enhancers and involvement of 9G8 as a trans-acting factor. J. Biol. Chem. 276, 7850–7858 (2001).
Spellman, R., Llorian, M. & Smith, C. W. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell 27, 420–434 (2007).
Makeyev, E. V., Zhang, J., Carrasco, M. A. & Maniatis, T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 (2007).
Lareau, L. F., Inada, M., Green, R. E., Wengrod, J. C. & Brenner, S. E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446, 926–929 (2007).
Morrison, M., Harris, K. S. & Roth, M. B. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 94, 9782–9785 (1997).
Kalyna, M., Lopato, S. & Barta, A. Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell 14, 3565–3577 (2003).
Kalyna, M., Lopato, S., Voronin, V. & Barta, A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 34, 4395–4405 (2006).
Kumar, S. & Lopez, A. J. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J. 24, 2646–2655 (2005).
Pan, Q. et al. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 20, 153–158 (2006).
Rehwinkel, J., Raes, J. & Izaurralde, E. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem. Sci. 31, 639–646 (2006).
Martens, J. A., Wu, P. Y. & Winston, F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19, 2695–2704 (2005).
Thompson, D. M. & Parker, R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol. Cell. Biol. 27, 92–101 (2007).
Gaba, A., Jacobson, A. & Sachs, M. S. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20, 449–460 (2005).
Pain, V. M. Translational control during amino acid starvation. Biochimie 76, 718–728 (1994).
Gardner, L. B. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol. Cell. Biol. 28, 3729–3741 (2008).
Lew, J. E., Enomoto, S. & Berman, J. Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 18, 6121–6130 (1998).
Dahlseid, J. N. et al. mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae. Eukaryotic Cell 2, 134–142 (2003).
Enomoto, S., Glowczewski, L., Lew-Smith, J. & Berman, J. G. Telomere cap components influence the rate of senescence in telomerase-deficient yeast cells. Mol. Cell. Biol. 24, 837–845 (2004).
Reichenbach, P. et al. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 13, 568–574 (2003). This paper and reference 116 implicate SMG6 as a telomerase-associated factor that controls telomere length.
Keene, J. D. RNA regulons: coordination of post-transcriptional events. Nature Rev. Genet. 8, 533–543 (2007).
St Johnston, D. The beginning of the end. EMBO J. 20, 6169–6179 (2001).
Roegiers, F. & Jan, Y. N. Staufen: a common component of mRNA transport in oocytes and neurons? Trends Cell Biol. 10, 220–224 (2000).
Mouland, A. J. et al. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 74, 5441–5451 (2000).
Chatel-Chaix, L. et al. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 24, 2637–2648 (2004).
Kim, Y. K., Furic, L., Desgroseillers, L. & Maquat, L. E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120, 195–208 (2005). This is the first demonstration that UPF1 can be recruited to particular mRNAs by a mechanism other than NMD to trigger mRNA decay when translation terminates normally.
Kim, Y. K. et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 26, 2670–2681 (2007).
Furic, L., Maher-Laporte, M. & DesGroseillers, L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA 14, 324–335 (2008).
Monshausen, M., Gehring, N. H. & Kosik, K. S. The mammalian RNA-binding protein Staufen2 links nuclear and cytoplasmic RNA processing pathways in neurons. Neuromolecular Med. 6, 127–144 (2004).
Kaygun, H. & Marzluff, W. F. in Nonsense-Mediated mRNA Decay (ed. L. E. Maquat) 237–253 (Landes Bioscience, Georgetown, 2006).
Kaygun, H. & Marzluff, W. F. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol. Cell. Biol. 25, 6879–6888 (2005). This is an important demonstration that SLBP recruits UPF1 to the 3′ UTR of replication-dependent histone mRNAs to elicit histone mRNA decay at the end of the S phase of the cell cycle.
Kaygun, H. & Marzluff, W. F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nature Struct. Mol. Biol. 12, 794–800 (2005).
Muller, B., Blackburn, J., Feijoo, C., Zhao, X. & Smythe, C. DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J. Cell Biol. 179, 1385–1398 (2007).
Azzalin, C. M. & Lingner, J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 16, 433–439 (2006). A significant piece of work demonstrating that UPF1 is required for human cells to progress completely through S phase and that ATR-mediated UPF1 phosphorylation promotes UPF1 binding to chromatin.
van Heemst, D., den Reijer, P. M. & Westendorp, R. G. Ageing or cancer: a review on the role of caretakers and gatekeepers. Eur. J. Cancer 43, 2144–2152 (2007).
Shiloh, Y. ATM and related protein kinases: safeguarding genome integrity. Nature Rev. Cancer 3, 155–168 (2003).
Erker, L. et al. Cancer chemoprevention by the antioxidant tempol acts partially via the p53 tumor suppressor. Hum. Mol. Genet. 14, 1699–1708 (2005).
Gehen, S. C., Staversky, R. J., Bambara, R. A., Keng, P. C. & O'Reilly, M. A. hSMG-1 and ATM sequentially and independently regulate the G(1) checkpoint during oxidative stress. Oncogene 10 Mar 2008 (doi:10.1038/onc.2008.48).
Oliveira, V. et al. A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J. Biol. Chem. 283, 13174–13184 (2008).
Abraham, R. T. & Oliveira, V. in Nonsense-Mediated mRNA Decay (ed. L. E. Maquat) 215–229 (Landes Bioscience, Georgetown, 2006).
Neben, K. et al. Distinct gene expression patterns associated with FLT3- and NRAS-activating mutations in acute myeloid leukemia with normal karyotype. Oncogene 24, 1580–1588 (2005).
Pal, M., Ishigaki, Y., Nagy, E. & Maquat, L. E. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7, 5–15 (2001).
Carastro, L. M. et al. Identification of delta helicase as the bovine homolog of HUPF1: demonstration of an interaction with the third subunit of DNA polymerase delta. Nucleic Acids Res. 30, 2232–2243 (2002).
Cheung, A. L. & Deng, W. Telomere dysfunction, genome instability and cancer. Front. Biosci. 13, 2075–2090 (2008).
Snow, B. E. et al. Functional conservation of the telomerase protein Est1p in humans. Curr. Biol. 13, 698–704 (2003).
Redon, S., Reichenbach, P. & Lingner, J. Protein RNA and protein protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 35, 7011–7022 (2007).
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E. & Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 (2007). A landmark paper showing that mammalian telomeres are transcribed, and telomere integrity is regulated, by NMD factors.
Hock, J. et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 8, 1052–1060 (2007).
Ajamian, L. et al. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14, 914–927 (2008).
Kataoka, N., Diem, M. D., Kim, V. N., Yong, J. & Dreyfuss, G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 20, 6424–6433 (2001).
Dostie, J. & Dreyfuss, G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12, 1060–1067 (2002).
Chan, C. C. et al. eIF4A3 is a novel component of the exon junction complex. RNA 10, 200–209 (2004).
Degot, S. et al. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J. Biol. Chem. 279, 33702–33715 (2004).
Baguet, A. et al. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J. Cell. Sci. 120, 2774–2784 (2007).
Palacios, I. M., Gatfield, D., St Johnston, D. & Izaurralde, E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427, 753–757 (2004).
Denning, G., Jamieson, L., Maquat, L. E., Thompson, E. A. & Fields, A. P. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J. Biol. Chem. 276, 22709–22714 (2001).
Fukuhara, N. et al. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 17, 537–547 (2005).
Kadlec, J., Guilligay, D., Ravelli, R. B. & Cusack, S. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 12, 1817–1824 (2006).
Kadlec, J., Izaurralde, E. & Cusack, S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nature Struct. Mol. Biol. 11, 330–337 (2004).
Kim, V. N. et al. The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J. 20, 2062–2068 (2001).
Chiu, S. Y., Serin, G., Ohara, O. & Maquat, L. E. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9, 77–87 (2003).
Le Hir, H., Gatfield, D., Braun, I. C., Forler, D. & Izaurralde, E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2, 1119–1124 (2001).
Shibuya, T., Tange, T. O., Sonenberg, N. & Moore, M. J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nature Struct. Mol. Biol. 11, 346–351 (2004).
Ferraiuolo, M. A. et al. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl Acad. Sci. USA 101, 4118–4123 (2004).
Acknowledgements
We thank C. Woeller for reading the manuscript and help with its formatting and G. Chanfreau for conversations. The Maquat lab is supported by NIH R01 grants GM074593 and GM059514 to L.E.M.
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- Telomeres
-
Condensed repetitive DNA sequences at chromosomal ends in most eukaryotes that compensate for incomplete semi-conservative DNA replication by protecting against homologous recombination and non-homologous end joining, thereby conferring genomic stability.
- Oxidative stress
-
A disturbance in the normal redox state of a cell that is due to an imbalance between the production of reactive oxygen and either detoxification of the resulting reactive intermediates (for example, peroxides and free radicals) or repair of the consequential damage. Oxidative-stress damage can potentially occur to all components of the cell, including proteins, lipids and DNA.
- Phosphatidylinositol 3-kinase-related protein kinase
-
(PIKK). Examples include ATM, ATR, SMG1, DNA-PK and mTOR. PIKKs constitute a subfamily of serine and threonine kinases that resemble lipid kinase phosphatidylinositol 3-kinases (PI-3 kinases). PIKKs transduce signals in cell-growth and stress-response pathways.
- Histone
-
Protein component of chromatin that functions to regulate gene expression and is synthesized coordinately with DNA replication. Two each of the core histones H2A, H2B, H3 and H4 make up an octameric nucleosome, around which DNA winds. The linker histone H1 binds the nucleosome, locking the DNA into place.
- Premature termination codon
-
(PTC). UAA, UGA or UAG codon (that is, a nonsense codon, which generally does not encode an amino acid) within mRNA. PTCs are situated upstream of the normal termination codon and direct the premature termination of mRNA translation, which usually results in nonsense-mediated mRNA decay.
- Exon-junction complex
-
(EJC). Complex of proteins that is deposited ∼20–25 nucleotides upstream of the exon–exon junctions of newly synthesized spliced mRNAs. Despite its potentially ancient origin, the EJC functions in mammalian-cell and plant-cell NMD but not detectably in NMD in other organisms studied.
- RNA cap-binding protein
-
(CBP). Protein that binds the 7-methyl guanosine cap structure at the 5′ end of mRNAs. In mammalian cells, mRNA is bound first by the mostly nuclear but shuttling CBP heterodimer CBP80–CBP20. Subsequently, CBP80–CBP20 is replaced by eukaryotic translation initiation factor 4E (eIF4E), a state that typifies the bulk of cellular mRNA.
- NMD degradative activities
-
Decapping and 5′-to-3′ exonucleolytic activities as well as deadenylating and 3′-to-5′ exosome-mediated activities.
- Selenoprotein mRNA
-
One of a number of mRNAs, exemplified by glutathione peroxidase 1 mRNA, that harbour one or more UGA codons that direct the incorporation of the twenty-first amino acid, selenocysteine. Incorporation, which competes with translation termination, requires at least one cis-acting selenocysteine insertion element that resides in the mRNA 3′ UTR.
- Serine–arginine (SR)-rich protein
-
Member of an evolutionarily conserved family of essential pre-mRNA splicing factors in metazoans. Individual SR proteins have distinct but occasionally overlapping abilities to promote 5′ splice-site usage. They also function in post-transcriptional steps that occur after pre-mRNA splicing.
- Heterogeneous nuclear ribonucleoprotein (hnRNP) splicing factor
-
hnRNP proteins that, in metazoans, can either repress or enhance splicing by antagonizing or promoting splice-site selection. Cooperative interactions between pre-mRNA-bound hnRNP proteins might enhance splicing between specific pairs of splice sites while at the same time inhibiting other combinations of splice-site usage.
- Alternative splicing
-
Used to generate the multiple mRNAs that derive from at least 75% of human genes, often in a tissue-specific manner, as a means to generate more than one protein isoform per gene. An estimated one-third of alternatively spliced human mRNAs are targeted for NMD.
- Pre-mRNA splicing
-
Nuclear process whereby one or more introns, or intervening sequences, are removed from a primary gene transcript, or a pre-mRNA, and the resulting exons are coordinately ligated together to form mRNA.
- Upstream ORF
-
An open translational reading frame that exists upstream of a protein-encoding reading frame. Upstream ORFs are usually short and are often regulatory, lacking the potential to encode a functional protein. Translation termination at an upstream ORF can result in NMD.
- Telomerase
-
A reverse transcriptase that uses its constituent RNA as a template to add specific DNA repeats — TTAGGG in all vertebrates — to the 3′-extended ends of telomeres, which are shortened after each replication cycle.
Rights and permissions
About this article
Cite this article
Isken, O., Maquat, L. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet 9, 699–712 (2008). https://doi.org/10.1038/nrg2402
Issue Date:
DOI: https://doi.org/10.1038/nrg2402
This article is cited by
-
A novel long noncoding RNA Linc-ASEN represses cellular senescence through multileveled reduction of p21 expression
Cell Death & Differentiation (2020)
-
CSL controls telomere maintenance and genome stability in human dermal fibroblasts
Nature Communications (2019)
-
The coming-of-age of nucleocytoplasmic transport in motor neuron disease and neurodegeneration
Cellular and Molecular Life Sciences (2019)
-
The Post-GWAS Era: How to Validate the Contribution of Gene Variants in Lupus
Current Rheumatology Reports (2019)
-
The RNA surveillance proteins UPF1, UPF2 and SMG6 affect HIV-1 reactivation at a post-transcriptional level
Retrovirology (2018)