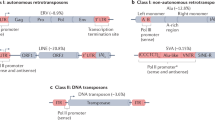
Abstract
The control and coordination of eukaryotic gene expression rely on transcriptional and post-transcriptional regulatory networks. Although progress has been made in mapping the components and deciphering the function of these networks, the mechanisms by which such intricate circuits originate and evolve remain poorly understood. Here I revisit and expand earlier models and propose that genomic repeats, and in particular transposable elements, have been a rich source of material for the assembly and tinkering of eukaryotic gene regulatory systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Britten, R. J. & Kohne, D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 161, 529–540 (1968).
Wicker, T. et al. A unified classification system for eukaryotic transposable elements. Nature Rev. Genet. 8, 973–982 (2007).
Brookfield, J. F. The ecology of the genome — mobile DNA elements and their hosts. Nature Rev. Genet. 6, 128–136 (2005).
Kidwell, M. G. & Lisch, D. R. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution Int. J. Org. Evolution 55, 1–24 (2001).
Feschotte, C. & Pritham, E. J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368 (2007).
Deininger, P. L., Moran, J. V., Batzer, M. A. & Kazazian, H. H. Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 13, 651–658 (2003).
Gould, S. J. & Vrba, E. S. Exaptation — a missing term in the science of form. Paleobiology 8, 4–15 (1983).
Brosius, J. Retroposons — seeds of evolution. Science 251, 753 (1991).
Britten, R. J. Cases of ancient mobile element DNA insertions that now affect gene regulation. Mol. Phylogenet. Evol. 5, 13–17 (1996).
Miller, W. J., McDonald, J. F., Nouaud, D. & Anxolabehere, D. Molecular domestication — more than a sporadic episode in evolution. Genetica 107, 197–207 (1999).
Silva, J. C., Shabalina, S. A., Harris, D. G., Spouge, J. L. & Kondrashovi, A. S. Conserved fragments of transposable elements in intergenic regions: evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet. Res. 82, 1–18 (2003).
Lowe, C. B., Bejerano, G. & Haussler, D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc. Natl Acad. Sci. USA 104, 8005–8010 (2007).
Mikkelsen, T. S. et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447, 167–177 (2007).
Bejerano, G., Haussler, D. & Blanchette, M. Into the heart of darkness: large-scale clustering of human non-coding DNA. Bioinformatics 20 (Suppl. 1), i40–i48 (2004).
Xie, X., Kamal, M. & Lander, E. S. A family of conserved noncoding elements derived from an ancient transposable element. Proc. Natl Acad. Sci. USA 103, 11659–11664 (2006).
Bejerano, G. et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441, 87–90 (2006).
Kamal, M., Xie, X. & Lander, E. S. A large family of ancient repeat elements in the human genome is under strong selection. Proc. Natl Acad. Sci. USA 103, 2740–2745 (2006).
Nishihara, H., Smit, A. F. & Okada, N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 16, 864–874 (2006).
Santangelo, A. M. et al. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 3, 1813–1826 (2007).
Maside, X., Bartolome, C. & Charlesworth, B. S-element insertions are associated with the evolution of the HSP70 genes in Drosophila melanogaster. Curr. Biol. 12, 1686–1691 (2002).
Schlenke, T. A. & Begun, D. J. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl Acad. Sci. USA 101, 1626–1631 (2004).
Chung, H. et al. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175, 1071–1077 (2007).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Inada, D. C. et al. Conserved noncoding sequences in the grasses. Genome Res. 13, 2030–2041 (2003).
Brosius, J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica 118, 99–116 (2003).
Marino-Ramirez, L., Lewis, K. C., Landsman, D. & Jordan, I. K. Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet. Genome Res. 110, 333–341 (2005).
Jordan, I. K., Rogozin, I. B., Glazko, G. V. & Koonin, E. V. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19, 68–72 (2003).
van de Lagemaat, L. N., Landry, J. R., Mager, D. L. & Medstrand, P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 19, 530–536 (2003).
Marino-Ramirez, L. & Jordan, I. K. Transposable element derived DNaseI-hypersensitive sites in the human genome. Biol. Direct 1, 20 (2006).
Gentles, A. J. et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 17, 992–1004 (2007).
Ni, J. Z. et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 21, 708–718 (2007).
Wray, G. A. et al. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20, 1377–1419 (2003).
Hambor, J. E., Mennone, J., Coon, M. E., Hanke, J. H. & Kavathas, P. Identification and characterization of an Alu-containing, T-cell-specific enhancer located in the last intron of the human CD8 alpha gene. Mol.Cell Biol. 13, 7056–7070 (1993).
Zhou, Y. H., Zheng, J. B., Gu, X., Saunders, G. F. & Yung, W. K. Novel PAX6 binding sites in the human genome and the role of repetitive elements in the evolution of gene regulation. Genome Res. 12, 1716–1722 (2002).
Medstrand, P. et al. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet. Genome Res. 110, 342–352 (2005).
Thornburg, B. G., Gotea, V. & Makalowski, W. Transposable elements as a significant source of transcription regulating signals. Gene 365, 104–110 (2006).
Polak, P. & Domany, E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics 7, 133 (2006).
Johnson, R. et al. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 34, 3862–3877 (2006).
Wang, T. et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl Acad. Sci. USA 104, 18613–18618 (2007).
Wessler, S. R., Bureau, T. E. & White, S. E. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5, 814–821 (1995).
Ferrigno, O. et al. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nature Genet. 28, 77–81 (2001).
Romanish, M. T., Lock, W. M., van de Lagemaat, L. N., Dunn, C. A. & Mager, D. L. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 3, e10 (2007).
Borchert, G. M., Lanier, W. & Davidson, B. L. RNA polymerase III transcribes human microRNAs. Nature Struct. Mol. Biol. 13, 1097–1101 (2006).
Britten, R. J. & Davidson, E. H. Gene regulation for higher cells: a theory. Science 165, 349–357 (1969).
Britten, R. J. & Davidson, E. H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 46, 111–138 (1971).
Peaston, A. E. et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606 (2004).
Bringaud, F. et al. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 3, 1291–1307 (2007).
Wilkins, A. S. The Evolution of Developmental Pathways (Sinauer, Sunderland, Massachusetts, 2002).
Davidson, E. H. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution (Academic, New York, 2006).
Mattick, J. S. A new paradigm for developmental biology. J. Exp. Biol. 210, 1526–1547 (2007).
Chen, K. & Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nature Rev. Genet. 8, 93–103 (2007).
Grewal, S. I. & Jia, S. Heterochromatin revisited. Nature Rev. Genet. 8, 35–46 (2007).
Slotkin, R. K. & Martienssen, R. Transposable elements and the epigenetic regulation of the genome. Nature Rev. Genet. 8, 272–285 (2007).
Aravin, A. A., Hannon, G. J. & Brennecke, J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318, 761–764 (2007).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 5, 522–531 (2004).
Smalheiser, N. R. & Torvik, V. I. Mammalian microRNAs derived from genomic repeats. Trends Genet. 21, 322–326 (2005).
Piriyapongsa, J., Marino-Ramirez, L. & Jordan, I. K. Origin and evolution of human microRNAs from transposable elements. Genetics 176, 1323–1337 (2007).
Smalheiser, N. R. & Torvik, V. I. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 22, 532–536 (2006).
Feschotte, C., Jiang, N. & Wessler, S. R. Plant transposable elements: where genetics meets genomics. Nature Rev. Genet. 3, 329–341 (2002).
Sijen, T. & Plasterk, R. H. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426, 310–314 (2003).
Piriyapongsa, J. & Jordan, I. K. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE 2, e203 (2007).
Levine, M. & Tjian, R. Transcription regulation and animal diversity. Nature 424, 147–151 (2003).
Volff, J. N. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28, 913–922 (2006).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Pace, J. K. & Feschotte, C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 17, 422–432 (2007).
Zdobnov, E. M., Campillos, M., Harrington, E. D., Torrents, D. & Bork, P. Protein coding potential of retroviruses and other transposable elements in vertebrate genomes. Nucleic Acids Res. 33, 946–954 (2005).
Casola, C., Lawing, A. M., Betran, E. & Feschotte, C. PIF-like transposons are common in Drosophila and have been repeatedly domesticated to generate new host genes. Mol. Biol. Evol. 24, 1872–1888 (2007).
Campillos, M., Doerks, T., Shah, P. K. & Bork, P. Computational characterization of multiple gag-like human proteins. Trends Genet. 22, 585–589 (2006).
Muehlbauer, G. J. et al. A hAT superfamily transposase recruited by the cereal grass genome. Mol. Genet. Genomics 275, 553–563 (2006).
Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. Mobile DNA II, (American Society for Microbiology, Washington, 2002).
Makarova, K. S., Aravind, L. & Koonin, E. V. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 27, 384–386 (2002).
Ros, F. & Kunze, R. Regulation of activator/dissociation transposition by replication and DNA methylation. Genetics 157, 1723–1733 (2001).
Aravind, L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem. Sci. 25, 421–423 (2000).
Siegmund, T. & Lehmann, M. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev. Genes Evol. 212, 152–157 (2002).
Roussigne, M. et al. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem. Sci. 28, 66–69 (2003).
Kapitonov, V. V. & Jurka, J. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 23, 311–324 (2004).
Babu, M. M., Iyer, L. M., Balaji, S. & Aravind, L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 34, 6505–6520 (2006).
Tudor, M., Lobocka, M., Goodwell, M., Pettitt, J. & O'Hare, K. The pogo transposable element family of Drosophila melanogaster. Mol. Gen. Genet. 232, 126–134 (1992).
Franz, G., Loukeris, T. G., Dialektaki, G., Thompson, C. R. & Savakis, C. Mobile Minos elements from Drosophila hydei encode a two-exon transposase with similarity to the paired DNA-binding domain. Proc. Natl Acad. Sci. USA 91, 4746–4750 (1994).
Breitling, R. & Gerber, J. K. Origin of the paired domain. Dev. Genes Evol. 210, 644–650 (2000).
Quesneville, H., Nouaud, D. & Anxolabehere, D. Recurrent recruitment of the THAP DNA-binding domain and molecular domestication of the P-transposable element. Mol. Biol. Evol. 22, 741–746 (2005).
Casola, C., Hucks, D. & Feschotte, C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol., 25, 29–41 (2008).
Piriyapongsa, J., Rutledge, M. T., Patel, S., Borodovsky, M. & Jordan, I. K. Evaluating the protein coding potential of exonized transposable element sequences. Biol. Direct 2, 31 (2007).
Cowan, R. K., Hoen, D. R., Schoen, D. J. & Bureau, T. E. MUSTANG is a novel family of domesticated transposase genes found in diverse angiosperms. Mol. Biol. Evol. 22, 2084–2089 (2005).
Lin, R. et al. Transposase-derived transcription factors regulate light signalling in Arabidopsis. Science 318, 1302–1305 (2007).
Cordaux, R., Udit, S., Batzer, M. A. & Feschotte, C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl Acad. Sci. USA 103, 8101–8106 (2006).
Liu, D. et al. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell Biol. 27, 1125–1132 (2007).
Miskey, C. et al. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell Biol. 27, 4589–4600 (2007).
Pathak, R. U., Rangaraj, N., Kallappagoudar, S., Mishra, K. & Mishra, R. K. Boundary element-associated factor 32B connects chromatin domains to the nuclear matrix. Mol. Cell Biol. 27, 4796–4806 (2007).
Cam, H. P., Noma, K. I., Ebina, H., Levin, H. L. & Grewal, S. I. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451, 431–436 (2008).
Hudson, M. E., Lisch, D. R. & Quail, P. H. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34, 453–471 (2003).
Raizada, M. N., Brewer, K. V. & Walbot, V. A maize MuDR transposon promoter shows limited autoregulation. Mol. Genet. Genomics 265, 82–94 (2001).
Cui, H. & Fedoroff, N. V. Inducible DNA demethylation mediated by the maize Suppressor-mutator transposon-encoded TnpA protein. Plant Cell 14, 2883–2899 (2002).
Atkinson, P. W., Warren, W. D. & O'Brochta, D. A. The hobo transposable element of Drosophila can be cross-mobilized in houseflies and excises like the Ac element of maize. Proc. Natl Acad. Sci. USA 90, 9693–9697 (1993).
Rezsohazy, R., van Luenen, H. G. A. M., Durbin, R. M. & Plasterk, R. H. A. Tc7, a Tc1-hitch hiking transposon in Caenorhabditis elegans. Nucleic Acids Res. 25, 4048–4054 (1997).
Lampe, D. J., Walden, K. K. & Robertson, H. M. Loss of transposase–DNA interaction may underlie the divergence of mariner family transposable elements and the ability of more than one mariner to occupy the same genome. Mol. Biol. Evol. 18, 954–961 (2001).
Feschotte, C., Osterlund, M. T., Peeler, R. & Wessler, S. R. DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res. 33, 2153–2165 (2005).
Wallace, M. R. et al. A de novo Alu insertion results in neurofibromatosis type 1. Nature 353, 864–866 (1991).
Girard, L. & Freeling, M. Regulatory changes as a consequence of transposon insertion. Dev. Genet. 25, 291–296 (1999).
Simons, C., Pheasant, M., Makunin, I. V. & Mattick, J. S. Transposon-free regions in mammalian genomes. Genome Res. 16, 164–172 (2006).
Lerman, D. N. & Feder, M. E. Naturally occurring transposable elements disrupt hsp70 promoter function in Drosophila melanogaster. Mol. Biol. Evol. 22, 776–783 (2005).
Walser, J. C., Chen, B. & Feder, M. E. Heat-shock promoters: targets for evolution by P transposable elements in Drosophila. PLoS Genet. 2, e165 (2006).
Ackerman, H., Udalova, I., Hull, J. & Kwiatkowski, D. Evolution of a polymorphic regulatory element in interferon-gamma through transposition and mutation. Mol. Biol. Evol. 19, 884–890 (2002).
Martin, C. & Lister, C. Genome juggling by transposons: Tam3-induced rearrangements in Antirrhinum majus. Dev. Genet. 10, 438–451 (1989).
Koga, A., Iida, A., Hori, H., Shimada, A. & Shima, A. Vertebrate DNA transposon as a natural mutator: the medaka fish Tol2 element contributes to genetic variation without recognizable traces. Mol. Biol. Evol. 23, 1414–1419 (2006).
Acknowledgements
I owe many thanks to C. Casola, J-M. Deragon, J. Fondon, I. K. Jordan, A. Pires da Silva, E. Pritham and D. Voytas for discussions and comments during the preparation of this article. I also thank the two anonymous reviewers for their constructive comments and useful suggestions. The author apologizes to many colleagues whose relevant work and original articles could not be cited owing to space limitations. Research in the C. F. laboratory is supported by grants from the US National Institutes of Health.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Feschotte, C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9, 397–405 (2008). https://doi.org/10.1038/nrg2337
Issue Date:
DOI: https://doi.org/10.1038/nrg2337
This article is cited by
-
Cell-type differential targeting of SETDB1 prevents aberrant CTCF binding, chromatin looping, and cis-regulatory interactions
Nature Communications (2024)
-
Quasispecies productivity
The Science of Nature (2024)
-
Symbionts of Ciliates and Ciliates as Symbionts
Indian Journal of Microbiology (2024)
-
Transposable elements as essential elements in the control of gene expression
Mobile DNA (2023)
-
Identification of transcriptionally active transposons in Barley
BMC Genomic Data (2023)