Key Points
-
There have been many attempts to classify proteins into groups of related molecules in terms of their primary, secondary and tertiary structure. An overview of these classification schemes is presented.
-
Structural classifications of proteins rely on similarity or metrics and the subsequent clustering of related molecules, or protein families, into groups.
-
Some schemes are manually curated whereas others are automatically derived. Although the quality of manual curations might generally be higher, they do not scale well with the increasing amount of protein sequence and structure information.
-
The coverage of known protein-sequence space is not homogeneous across these schemes. We argue that more comparative analyses of these schemes are required to derive a unified structural classification and to understand their similarities and differences.
-
More recently, functional classifications of proteins have also appeared, ranging from enzyme/reaction classifications to functional roles or cellular localization. These classifications rely heavily on functional genomics experiments and sequence annotations.
-
In principle, structural and functional classifications of proteins are independent of one another, and their relationships will further our understanding of the cellular roles of protein families.
-
Ultimately, a unified scheme for both structure and function should rely on a natural classification that takes into account both the phylogeny and biochemistry of the different protein types in nature.
Abstract
We examine the structural and functional classifications of the protein universe, providing an overview of the existing classification schemes, their features and inter-relationships. We argue that a unified scheme should be based on a natural classification approach and that more comparative analyses of the present schemes are required both to understand their limitations and to help delimit the number of known protein folds and their corresponding functional roles in cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ridley, M. in Philosophy of Biology (ed. Ruse, M.) 167–179 (Macmillan Publishing Co., New York, 1989).
Asimov, I. A Short History of Biology (Thomas Nelson & Sons Ltd., London, 1964).
Eisenberg, D., Marcotte, E. M., Xenarios, I. & Yeates, T. O. Protein function in the post-genomic era. Nature 405, 823–826 (2000).
Swindells, M. B., Orengo, C. A., Jones, D. T., Hutchinson, E. G. & Thornton, J. M. Contemporary approaches to protein structure classification. Bioessays 20, 884–891 (1998).
Heger, A. & Holm, L. Towards a covering set of protein family profiles. Prog. Biophys. Mol. Biol. 73, 321–337 (2000). A comprehensive analysis of strategies and resources for protein-sequence clustering and protein-family identification.
Liu, J. & Rost, B. Domains, motifs and clusters in the protein universe. Curr. Opin. Chem. Biol. 7, 5–11 (2003). An overview of present methods for protein-sequence clustering.
Murzin, A. G., Brenner, S. E., Hubbart, T. & Chothia, C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540 (1995).
Lo Conte, L., Brenner, S. E., Hubbard, T. J., Chothia, C. & Murzin, A. G. SCOP database in 2002: refinements accommodate structural genomics. Nucl. Acids Res. 30, 264–267 (2002).
Orengo, C. A. et al. CATH- a hierarchic classification of protein domain structures. Structure 5, 1093–1108 (1997).
Pearl, F. M. et al. The CATH database: an extended protein family resource for structural and functional genomics. Nucl. Acids Res. 31, 452–455 (2003).
Holm, L., Ouzounis, C., Sander, C., Tuparev, G. & Vriend, G. A database of protein structure families with common folding motifs. Protein Sci. 1, 1691–1698 (1992).
Holm, L. & Sander, C. Touring protein fold space with Dali/FSSP. Nucl. Acids Res. 26, 316–319 (1998).
Orengo, C. A. & Taylor, W. R. SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol. 266, 617–635 (1996).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Holm, L. & Sander, C. Mapping the protein universe. Science 273, 595–602 (1996).
Brenner, S. E., Chothia, C. & Hubbard, T. J. Population statistics of protein structures: lessons from structural classifications. Curr. Opin. Struct. Biol. 7, 369–376 (1997).
Burley, S. K. & Bonanno, J. B. Structuring the universe of proteins. Ann. Rev. Genomics Hum. Genet. 3, 243–262 (2002).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Sander, C. & Schneider, R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins 9, 56–68 (1991).
Dodge, C., Schneider, R. & Sander, C. The HSSP database of protein structure-sequence alignments and family profiles. Nucl. Acids Res. 26, 313–315 (1998).
Sonnhammer, E. L., Eddy, S. R. & Durbin, R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28, 405–420 (1997).
Bateman, A. et al. The Pfam protein families database. Nucl. Acids Res. 30, 276–280 (2002).
Attwood, T. K., Beck, M. E., Bleasby, A. J. & Parry-Smith, D. J. PRINTS — a database of protein motif fingerprints. Nucl. Acids Res. 22, 3590–3596 (1994).
Attwood, T. K. et al. PRINTS and its automatic supplement, prePRINTS. Nucl. Acids Res. 31, 400–402 (2003).
Schultz, J., Milpets, F., Bork, P. & Ponting, C. P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA 95, 5857–5864 (1998).
Letunic, I. et al. Recent improvements to the SMART domain-based sequence annotation resource. Nucl. Acids Res. 30, 242–244 (2002).
Bairoch, A. PROSITE: a dictionary of sites and patterns in proteins. Nucl. Acids Res. 19, 2241–2245 (1991).
Falquet, L. et al. The PROSITE database, its status in 2002. Nucl. Acids Res. 30, 235–238 (2002).
Haft, D. H. et al. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucl. Acids Res. 29, 41–43 (2001).
Haft, D. H., Selengut, J. D. & White, O. The TIGRFAMs database of protein families. Nucl. Acids Res. 31, 371–373 (2003).
Corpet, F., Gouzy, J. & Kahn, D. The ProDom database of protein domain families. Nucl. Acids Res. 26, 323–326 (1998).
Corpet, F., Servant, F., Gouzy, J. & Kahn, D. ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucl. Acids Res. 28, 267–269 (2000).
Henikoff, S. & Henikoff, J. G. Automated assembly of protein blocks for database searching. Nucl. Acids Res. 19, 6565–65672 (1991).
Henikoff, S., Henikoff, J. G. & Pietrokovski, S. Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics 15, 471–479 (1999).
Nevill-Maning, C. G., Wu, T. D. & Brutlag, D. L. Highly specific protein sequence motifs for genome analysis. Proc. Natl Acad. Sci. USA 95, 5865–5871 (1998).
Huang, J. Y. & Brutlag, D. L. The EMOTIF database. Nucl. Acids Res. 29, 202–204 (2001).
Rigoutsos, I., Huynh, T., Floratos, A., Parida, L. & Platt, D. Dictionary-driven protein annotation. Nucl. Acids Res. 30, 3901–3916 (2002).
Krause, A., Haas, S. A., Coward, E. & Vingron, M. SYSTERS, GeneNet, SpliceNest: exploring sequence space from genome to protein. Nucl. Acids Res. 30, 299–300 (2002).
Kriventseva, E. V., Fleischmann, W., Zdobnov, E. M. & Apweiler, R. CluSTr: a database of clusters of SWISS-PROT+TrEMBL proteins. Nucl. Acids Res. 29, 33–36 (2001).
Kriventseva, E. V., Servant, F. & Apweiler, R. Improvements to CluSTr: the database of SWISS-PROT+TrEMBL protein clusters. Nucl. Acids Res. 31, 388–389 (2003).
Tatusov, R. L., Koonin, E. V. & Lipman, D. J. A genomic perspective on protein families. Science 278, 631–637 (1997).
Wheeler, D. L. et al. Database resources of the National Center for Biotechnology Information. Nucl. Acids Res. 31, 28–33 (2003).
Yona, G., Linial, N. & Linial, M. ProtoMap: automatic classification of protein sequences, a hierarchy of protein families, and local maps of the protein space. Proteins 37, 360–378 (1999).
Yona, G., Linial, N. & Linial, M. ProtoMap: automatic classification of protein sequences and hierarchy of protein families. Nucl. Acids Res. 28, 49–55 (2000).
Silverstein, K. A., Shoop, E., Johnson, J. E. & Retzel, E. F. MetaFam: a unified classification of protein families. I. Overview and statistics. Bioinformatics 17, 249–261 (2001).
Shoop, E., Silverstein, K. A., Johnson, J. E. & Retzel, E. F. MetaFam: a unified classification of protein families. II. Schema and query capabilities. Bioinformatics 17, 262–271 (2001).
Enright, A. J., Kunin, V. & Ouzounis, C. A. Protein families and TRIBEs in genome sequence space. Nucl. Acids Res. (in the press).
Mulder, N. J. et al. The InterPro database, 2003 brings increased coverage and new features. Nucl. Acids Res. 31, 315–318 (2003).
Rigoutsos, I. & Floratos, A. Combinatorial pattern discovery in biological sequences: the TEIRESIAS algorithm. Bioinformatics 14, 55–67 (1998).
Enright, A. J., van Dongen, S. & Ouzounis, C. A. An efficient algorithm for large-scale detection of protein families. Nucl. Acids Res. 30, 1575–1584 (2002).
Bairoch, A. The ENZYME data bank. Nucl. Acids Res. 22, 3626–3627 (1993).
Bairoch, A. The ENZYME database in 2000. Nucl. Acids Res. 28, 304–305 (2000).
Garrels, J. I. YPD — a database for the proteins of Saccharomyces cerevisiae. Nucl. Acids Res. 24, 46–49 (1996).
Hodges, P. E., McKee, A. H., Davis, B. P., Payne, W. E. & Garrels, J. I. The Yeast Proteome Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucl. Acids Res. 27, 69–73 (1999).
Cherry, J. M. et al. SGD: Saccharomyces Genome Database. Nucl. Acids Res. 26, 73–79 (1998).
Dwight, S. S. et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucl. Acids Res. 30, 69–72 (2002).
Mewes, H. W. et al. MIPS: a database for genomes and protein sequences. Nucl. Acids Res. 27, 44–48 (1999).
Mewes, H. W. et al. MIPS: a database for genomes and protein sequences. Nucl. Acids Res. 30, 31–34 (2002).
Overbeek, R. et al. WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucl. Acids Res. 28, 123–125 (2000).
Snel, B., Lehmann, G., Bork, P. & Huynen, M. A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucl. Acids Res. 28, 3442–3444 (2000).
von Mering, C. et al. STRING: a database of predicted functional associations between proteins. Nucl. Acids Res. 31, 258–261 (2003).
Marcotte, E. M. et al. Detecting protein function and protein–protein interactions from genome sequences. Science 285, 751–753 (1999).
Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA 96, 4285–4288 (1999). This paper introduces the concept of phylogenetic profiles, and the idea that similar phylogenetic profiles indicate functional association between genes.
Enright, A. J. & Ouzounis, C. A. Functional associations of proteins in entire genomes via exhaustive detection of gene fusion. Genome Biol. 2, 0031–0037 (2001).
Enright, A. J., Iliopoulos, I., Kyrpides, N. C. & Ouzounis, C. A. Protein interaction maps for complete genomes based on gene fusion events. Nature 402, 86–90 (1999).
Yanai, I., Derti, A. & DeLici, C. Genes linked by fusion events are generally of the same functional category: a systematic analysis of 30 microbial genomes. Proc. Natl Acad. Sci. USA 98, 7940–7945 (2001). This paper is a 'proof of principle' that gene-fusion events can be used to infer functional associations, as proposed in references 63 and 65.
Mellor, J. C., Yanai, I., Clodfelter, K. H., Mintseris, J. & DeLisi, C. Predictome: a database of putative functional links between proteins. Nucl. Acids Res. 30, 306–309 (2002).
Riley, M. Functions of the gene products of Escherichia coli. Microbiol. Rev. 57, 862–952 (1993). The original comprehensive functional-classification scheme, developed for the gene products of the E. coli genome.
Serres, M. H. & Riley, M. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb. Comp. Genomics 5, 205–222 (2000).
Andrade, M. A. et al. Automated genome sequence analysis and annotation. Bioinformatics 15, 391–412 (1999).
Tamames, J., Ouzounis, C., Casari, G., Sander, C. & Valencia, A. EUCLID: automatic classification of proteins in functional classes by their database annotations. Bioinformatics 14, 542–543 (1998).
Ashburner, M. A. et al. Gene ontology: tool for the unification of biology. Nature Genet. 25, 25–29 (2000). This paper describes the development of a dynamic controlled vocabulary for the functional annotation of eukaryotic gene products.
Karp, P. D., Riley, M., Paley, S. M. & Pellegrini-Toole, A. EcoCyc: an encyclopedia of Escherichia coli genes and metabolism. Nucl. Acids Res. 24, 32–39 (1996).
Karp, P. D., Ouzounis, C. & Paley, S. HinCyc: a knowledge base of the complete genome and metabolic pathways of H. influenzae. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4, 116–124 (1996).
Karp, P. D., Riley, M., Paley, S. M. & Pellegrini-Toole, A. The MetaCyc database. Nucl. Acids Res. 30, 59–61 (2002).
Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. The KEGG databases at GenomeNet. Nucl. Acids Res. 30, 42–46 (2002).
Ogata, H. et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl. Acids Res. 27, 29–34 (1999).
Xenarios, I. et al. DIP: the database of interacting proteins. Nucl. Acids Res. 28, 289–291 (2000).
Xenarios, I. et al. DIP, the Database of Interacting Protiens: a research tool for studying cellular networks of protein interactions. Nucl. Acids Res. 30, 303–305 (2002).
Habeler, G. et al. YPL.db: the Yeast Protein Localization database. Nucl. Acids Res. 30, 80–83 (2002).
Kumar, A. et al. TRIPLES: a database of gene function in Saccharomyces cerevisiae. Nucl. Acids Res. 28, 81–84 (2000).
Kumar, A. et al. The TRIPLES database: a community resource for yeast molecular biology. Nucl. Acids Res. 30, 73–75 (2002).
Zanzoni, A. et al. MINT: a Molecular INTeraction database. FEBS Lett. 513, 135–140 (2002).
Bader, G. D. et al. BIND — the Biomolecular Interaction Network Database. Nucl. Acids Res. 29, 242–245 (2001).
Bader, G. D., Betel, D. & Hogue, C. W. BIND: the Biomolecular Interaction Network Database. Nucl. Acids Res. 31, 248–250 (2003).
Rain, J. C. et al. The protein–protein interaction map of Helicobacter pylori. Nature 409, 211–215 (2001). The only genome-wide protein-interaction map, so far, to be constructed for a prokaryote.
Gavin, A. C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002). This paper describes the identification of yeast-protein complexes using large-scale tandem-affinity purification coupled to mass spectrometry.
Bader, G. D. & Hogue, C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2 (2003).
Rison, S. C., Hodgman, T. C. & Thornton, J. M. Comparison of functional annotation schemes for genomes. Funct. Integr. Genomics 1, 56–69 (2000). An in-depth analysis and comparison of present functional classification schemes.
Iliopoulos, I. et al. Evaluation of annotation strategies using an entire genome sequence. Bioinformatics 19, 717–726 (2003).
Koonin, E. V., Wolf, Y. I. & Karev, G. P. The structure of the protein universe and genome evolution. Nature 420, 218–223 (2002).
von Mering, C. et al. Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417, 399–403 (2002).
Deane, C. M., Salwinski, L., Xenarios, I. & Eisenberg, D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell Proteomics 1, 349–356 (2002).
Mayr, E. Biological classification: toward a synthesis of opposing methodologies. Science 214, 510–516 (1981).
Jenssen, T. K., Laegreid, A., Komorowski, J. & Hovig, E. A literature network of human genes for high-throughput analysis of gene expression. Nature Genet. 28, 21–28 (2001). An automated analysis of the biomedical literature that identifies large-scale functional associations between thousands of human genes.
Stephens, R. S. et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759 (1998).
Fromont-Racine, M. et al. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 17, 95–110 (2000). This paper describes the first large-scale use of two-hybrid arrays to identify protein interactions in yeast.
Uetz, P. et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 (2000).
Acknowledgements
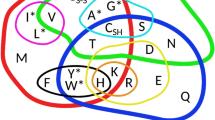
R.M.R.C. would like to acknowledge the Medical Research Council (UK) for support. J.P.L. would like to acknowledge the Foundation for Science and Technology (Portugal). We also thank C. Von Mering, I. Rigoutsos and colleagues at the European Bioinformatics Institute (EBI) for providing information on Figure 1.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
Further Information
Glossary
- METRIC
-
A criterion or set of criteria that are stated in quantifiable terms.
- DISTANCE-BASED HIERARCHICAL CLUSTERING
-
Clustering is the process of grouping objects on the basis of their similarity. Distance-based hierarchical clustering is used to construct a tree of nested clusters on the basis of the proximity (or distance) between data points.
- STRUCTURAL GENOMICS
-
Initiatives to solve the structures of proteins that are encoded in an entire genome by high-throughput methods.
- POSITION-WEIGHTED DYNAMIC PROGRAMMING
-
Dynamic programming is an algorithmic approach to solve sequential or multi-stage decision problems, such as finding optimal protein-sequence alignments. The position-weighted dynamic-programming method incorporates a matrix of substitution frequencies between amino acids, weighted by the degree of conservation of particular residues.
- SEED ALIGNMENTS
-
Hand-edited multiple sequence alignments that incorporate sequences that are described in the literature as belonging to the same family. From these seed alignments, hidden Markov models can be created that can in turn be used to search databases and identify new members of the family.
- HIDDEN MARKOV MODEL
-
(HMM). A pattern-recognition approach that is used in bioinformatics for DNA/protein feature detection and sequence comparison. HMMs are based on transition probabilities for discrete states. These probabilities are usually derived from training sets such as seed alignments.
- COMBINATORIAL PATTERN DISCOVERY
-
An approach that produces all patterns in any given data set in an efficient way that avoids the explicit enumeration of the entire pattern space.
- ORTHOLOGUES
-
Genes of common origin that have diverged through speciation rather than duplication. This term is sometimes ambiguously used to denote functionally equivalent genes that are of common origin in different organisms.
- NUCLEAR MAGNETIC RESSONANCE
-
(NMR). An analytical chemistry technique that is used to study molecular structure and dynamics, which explores spectrum differences that are caused by the differential alignment of atomic spins in the presence of a strong magnetic field.
- ENTROPY
-
A measure of the disorder or unavailability of energy within a closed system.
- ONTOGENY
-
The development and life cycle of a single organism.
- ONTOLOGY
-
An explicit formal specification of how to represent the objects, concepts and other entities within a domain of discourse, and the relationships among them. Ontologies are designed to create agreed vocabularies for exchanging information.
Rights and permissions
About this article
Cite this article
Ouzounis, C., Coulson, R., Enright, A. et al. Classification schemes for protein structure and function. Nat Rev Genet 4, 508–519 (2003). https://doi.org/10.1038/nrg1113
Issue Date:
DOI: https://doi.org/10.1038/nrg1113
This article is cited by
-
The coupling of taxonomy and function in microbiomes
Biology & Philosophy (2017)
-
Linking in domain-swapped protein dimers
Scientific Reports (2016)
-
Functional Genomics Evidence Unearths New Moonlighting Roles of Outer Ring Coat Nucleoporins
Scientific Reports (2014)
-
MESSA: MEta-Server for protein Sequence Analysis
BMC Biology (2012)
-
Self consistency grouping: a stringent clustering method
BMC Bioinformatics (2012)