Abstract
After an illustrious history as one of the primary tools that established the foundations of molecular biology, bacteriophage research is now undergoing a renaissance in which the primary focus is on the phages themselves rather than the molecular mechanisms that they explain. Studies of the evolution of phages and their role in natural ecosystems are flourishing. Practical questions, such as how to use phages to combat human diseases that are caused by bacteria, how to eradicate phage pests in the food industry and what role they have in the causation of human diseases, are receiving increased attention. Phages are also useful in the deeper exploration of basic molecular and biophysical questions.
Similar content being viewed by others
Main
Approximately 25 years after their discovery in 1915 (Ref. 1), the use of phages to answer fundamental questions about the mechanisms of inheritance began, taking advantage of the ease with which they could be manipulated2. Between the years 1940 and 1970, phage experiments laid much of the groundwork for the science of molecular biology. That science, once dubbed 'modern' as distinct from 'classical' biology, has long since been assimilated into the textbooks. However, recent research indicates that the historical contributions that phage studies have made to molecular biology might be overshadowed by their future influence on biology in general, and on genetics in particular. Phages have important roles in global ecology and bacterial pathogenicity. These emerging roles, together with the increasing number of recent publications on phage genomics, highlight the need for a timely assessment of phage biology and its bright future. Here, I summarize the studies of the molecular, ecological and evolutionary biology of phages that have led to this renaissance of interest, and define some crucial unanswered questions.
Background
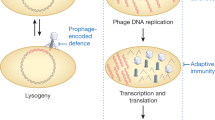
Bacteriophages (also known as phages) are viruses that infect bacteria. As with all viruses, phages are infectious particles that have at least two components: nucleic acid and protein. If a phage invades a susceptible bacterial cell, the phage (or at least its nucleic acid) enters the cell and triggers a cycle of phage production (Fig. 1). During this cycle, the cell is reprogrammed to become a phage factory in which the components of the biosynthetic apparatus (such as ribosomes and ATP generators) are diverted from their normal tasks in bacterial growth. The various pathways for reprogramming are initiated by phage-specified proteins, which are translated from phage mRNA that is made after infection.
The phage particle encounters and attaches to the cell surface by the tip of its tail and phage DNA enters, which leaves an empty protein shell attached to the outside of the cell. Next, the ends of the linear DNA molecule join to form a circle. The point of closure is called the cohesive site (cos). In some infected cells, the DNA is transcribed, translated and replicated. There are two pathways of replication: θ-form and rolling circle. Rolling-circle replication generates multigenomic tails of linear double-stranded DNA (dsDNA) from which DNA is drawn into preformed protein shells; tails are added and the cell lyses to liberate a crop of phage progeny. In other infected cells, phage development is repressed and phage DNA integrates into the bacterial chromosome. The resulting lysogenic cell can replicate indefinitely, but can be induced to return to the lytic cycle with the excision of phage DNA from the chromosome.
The temporal programme is ordered and regulated. Generally, nucleic-acid replication occurs first, followed by the synthesis of structural proteins of the phage particle. New phage particles are then assembled, and are later released from the cell. For most of the classically studied COLIPHAGES, release is effected by the disruption of the cell envelope and sudden lysis of the cell, the contents of which (including the new phage particles) spill out into the medium. The whole cycle can last ∼40 minutes and produce ∼100 new phages (Fig. 1).
This type of lytic cycle is the only mode of reproduction for many phages. Other TEMPERATE phages have an alternative life cycle in which a fraction of the infected cells go into the lytic cycle, whilst other cells survive infection and permanently harbour the phage genome in a quiescent form (PROPHAGE). Cells that harbour a prophage are known as LYSOGENIC. Certain agents (such as ultraviolet light) can induce lysogenic cells to re-enter the lytic cycle (Fig. 1).
One reason that phages have been useful in research is the ease of synchronizing the lytic cycle in a population of cells, so that the progression of molecular events can be measured over the whole culture. Synchronization might be achieved either by simultaneous infection or by the induction of phage development in lysogenic cells. A second reason is the ease with which mutations that affect specific stages of the cycle can be isolated and analysed.
Phages are as varied in size and form as the viruses that infect eukaryotes. Several formal classification schemes have been proposed, but none is widely useful or known to reflect phylogeny. Table 1 summarizes basic information on common model phages and illustrates some of the diversity in phage morphology and lifestyle.
Evolution
The phylogeny of viruses has eluded generations of virologists. Modern methods have redefined the questions rather than achieved definitive answers. In closely related groups, isolates can be arranged by the degree of similarity, but whether all viruses (or even all phages) constitute a MONOPHYLETIC GROUP remains uncertain.
The development of ideas, starting in the pre-genome era, is illustrated by work on phages and their relatives. In the 1950s, numerous temperate phages (LAMBDOIDS) were discovered that resembled the phage-λ morphologically and could recombine with it. Electron microscopy of HETERODUPLEXES, coupled with the results from genetic crosses, verified that the lambdoid phages have a common genetic map but show diverse specificities for properties such as integration, repression and host range. These studies also showed that any two lambdoid phages are similar in some segments of their genomes and dissimilar in others, with abrupt transitions from similarity to dissimilarity3.
Extensive DNA sequencing has confirmed this general picture. It has also shown that segments with altered specificity (such as repressor genes), although differing greatly in sequence, are nonetheless ORTHOLOGOUS. Comparing all the known lambdoid phages, there is little doubt that they have undergone extensive recombination in nature, mixing and matching heterologous specificity determinants. Sequencing also verifies the sharpness of most boundaries between segments of close similarity and those of extreme dissimilarity.
Hendrix4 offered the most comprehensive interpretation of these results. If different closely related phages, such as lambdoids, encounter one another in nature, they can recombine. The frequency of recombination depends on the degree of similarity of the two partners at the recombination points. So, between any two phages, shared segments of HOMOLOGY can serve as recombinational linkers, and even identities of oligonucleotide length can create preferred exchange points. None of this is new. However, breakage and joining sometimes occur even if there is no homology — which represents new ground — and over evolutionary time this happens often enough to generate the sharp boundaries between homology and non-homology that are observed in phages. Most such new junctions should be lethal or deleterious, but natural selection has preserved some benign combinations.
This raises the question of how the parental phages that give rise to a new junction came to be so different from one another. However, before addressing this issue, we leave the myopic consideration of lambdoid phages to look at what genome analysis implies about phages that are essentially unrelated, even those that come from highly diverse hosts.
On that level, the message is clear. Natural phage genomes are frequently chimeric in origin and contain genes that have been transferred over great evolutionary distances5. As with the genesis of new junctions among the lambdoid phages, the potential number of such transfers must greatly exceed what has passed through the sieve of natural selection. Among the more marked examples of molecular chimerism is phage N15. Whereas most of its phage genes are related to, and in the same order as, the genes of a lambdoid phage, its prophage form persists as a linear plasmid that includes sequences that are related to the F plasmid of Escherichia coli as well as to coliphages P1 and P4 (Ref. 6).
This widespread chimerism forces us to believe that phage genomes collectively constitute a pool that is subject to continual mixing. This pool includes groups, such as the lambdoid phages, in which mixing is even more frequent. However, some gene blocks that are found in lambdoid phages might have resided elsewhere for much of their history, and have diverged over long periods during which they were not subject to frequent recombination with other lambdoid phages. It should be noted that molecular clock estimates indicate that some allelic variants of λ-genes diverged long before the existence of E. coli, which is the host of λ, perhaps even as long ago as the branching of bacteria from archaea and eukaryotes4,7. There are many reasons to question the accuracy of dating by molecular clocks8. However, the antiquity of viral genes is also indicated by an independent line of evidence9. There are marked examples (such as adenoviruses and reoviruses) in which the morphology of the viral capsid is similar in a group that includes animal viruses, plant viruses and bacteriophages. These similarities extend from the shapes of individual proteins to the overall morphology of the virus particle (composed of several protein species) and seem unlikely to represent evolutionary convergence. This indicates a common ancestry for reoviruses and phage φ6, for example, although the RNA and protein sequences show no significant similarities and, therefore, must have diverged anciently, perhaps before the separation of the main domains of the living world.
One characteristic of a phage family such as the lambdoids, is a common genetic map. The main determinants for structural and regulatory elements in the phage genome are present in the same order throughout the family. As discussed previously, these elements have sometimes been replaced by homologues or ANALOGUES from other phages. However, at some time in the past these genomes must have come into being, probably by the merging of smaller genomes with more limited functions. So, the original lambdoid phage might have derived its replication genes from one source, its lysis genes from a second source and its structural genes from a third. Genes might have been added one at a time or in larger blocks3,4.
The exciting point is that such gene addition is not just a long ago fait accompli but is an active continuing process. The Hendrix group4 have identified segments of the genomes of lambdoid phages that seem to have been spliced into the phage genome recently. These segments (dubbed 'morons') typically include a single gene, which is often of unknown function, and generally differ in base composition from the rest of the phage genome. Each moron is present in some lambdoid phages and absent from others. The incorporation by phages of toxin genes and longer pathogenicity islands might be an example of the same kind of process (see later).
A complete understanding of the natural dynamics that underlie the recombinational origin of new phage types will require more specific information on the role of natural selection — not only the elimination of maladapted types (as mentioned earlier) but also active selection for the few superior variants. Further efforts in this direction are desirable, not just for their intrinsic interest but also as models for viruses that infect eukaryotic hosts; for example, it is well documented that members of the coronavirus group, which has attracted attention recently through the emergence of a deadly new human pathogen that causes severe acute respiratory syndrome (SARS), undergo frequent recombination in nature10,11.
Ecology
The long-held suspicion that phages are ubiquitous throughout the prokaryotic world has been confirmed as our knowledge has expanded to include more phages and phage-related sequences in prokaryotic genomes. The numerical abundance of phage-like particles in aquatic ecosystems generally exceeds that of prokaryotic cells and might represent >10% of the biomass of those cells. Furthermore, as documented in recent reviews12,13,14, there is increasing evidence to indicate that a large fraction of these visible particles comprises viable infectious phages.
At the base of the natural food chain are the photosynthetic microbes: green algae and cyanobacteria. For both of these groups, a substantial fraction of their biomass is liberated as dissolved organic material by viral infection. Heterotrophic consumers of dissolved organic matter are also frequently killed by viral infection. No account of the natural food web that omits bacteriophages is even close to being complete15,16.
Models for molecular biology
Molecular machines. The focus of biochemistry has advanced from chemical catalysis by individual proteins to mechanical processes that are carried out by molecular assemblages. Phage systems provide many instructive examples.
The forces that drive DNA injection during infection by the large-tailed DNA phages have long eluded direct identification. For many common phages, such as λ and T4, this remains the case. More progress has been made with phages, such as T7, which inject their DNA slowly. The initial step is the injection of T7 proteins from the phage particle to form a channel across the cell envelope that conducts DNA into the cell by a process that is apparently enzymatic17. Next, the DNA is pulled into the cell by transcription with an effectively stationary RNA polymerase (first host then phage polymerase) that reels DNA through itself18,19. The TYPE I RESTRICTION ENZYME Eco KI can replace RNA polymerase to reel in DNA20.
In assembling large double-stranded DNA (dsDNA) phages, viral DNA can be pulled into preformed heads by the portal protein that is located at one vertex of the icosahedron. Detailed studies of the physical mechanism were made on Bacillus phage φ29 (Ref. 21). The dodecameric portal protein rotates in the protein shell of the phage, in steps of 12 °, which reel in phage DNA and are powered by ATP hydrolysis (Fig. 2). The forces that act on individual DNA molecules that are being packaged into the heads of φ29 have been accurately measured, and show that the φ29 portal protein generates one of the strongest known molecular motors22.
An illustration of one cycle in the mechanism that rotates the connector and translates the DNA into the head. The view down the connector axis (top row) is towards the head, whereas the bottom row shows side views that correspond to panel b. Of the twelve subunits (A–L) of the connector, eleven are shown in green, and the one 'active' monomer is shown in red. The connector is represented as a set of small spheres at the narrow end and a set of larger spheres at the wide end, which are connected by a line that represents the central helical region. The plasmid RNA (pRNA)–ATPase complexes (I–V), which surround the narrow end, are shown by a set of four blue spheres and one red sphere. The DNA base that is aligned with the active connector monomer is also shown in red. a | The active pRNA–ATPase I interacts with the adjacent connector monomer (A), which in turn contacts the aligned DNA base. b | The narrow end of the connector has moved anti-clockwise by 12 °, to place the narrow end of monomer C opposite ATPase II, the next ATPase to be fired; this causes the connector to expand lengthwise by slightly altering the angle of the helices in the central domain (white arrow with asterisk). c | The wide end of the connector has followed the narrow end, as the connector relaxes and contracts (white arrow with two asterisks), thereby causing the DNA to be translated into the phage head. For the next cycle, ATPase II is activated, which causes the connector to be rotated by another 12 °, and so on. Figure reproduced with permission from Ref. 21.
As well as such motors, phages use numerous molecular machines. The insertion of phage DNA into the chromosome during lysogenization is a good example. Several molecules of λ-integrase form an assemblage (the INTASOME) that binds to a specific segment of phage DNA, recruits the bacterial insertion sites into the complex, then makes concerted single-strand cuts in the two DNAs. The strand cutting proceeds through the covalent joining of a 3′ phosphate to a tyrosine of the integrase protein. The terminal three bases of the free 5′ ends then rotate to exchange partners, followed by ligation with removal of the integrase protein from the 3′ ends to form a HOLLIDAY JUNCTION. The assemblage then undergoes a rearrangement (isomerization) and resolves the Holliday structure by cutting the two previously uncut strands on the other side of the junction, which allows the exchange of partners by the freed 5′ ends, followed by resealing23. The reverse reaction (excision from the chromosome) requires a second phage-specified protein (excisionase) and proceeds by an analogous mechanism, which is, however, not a simple chemical reversal of the integration reaction24.
Probably the most important recent breakthrough was the discovery that both initiation and resolution involve strand swapping of the free 5′ ends, as described earlier22. It was previously thought that Holliday junctions were formed by re-ligation at the cutting site, followed by branch migration to the resolution site (7-bps removed). Since then, important progress has been made on the nature of the initial DNA–protein interactions25,26 and the characterization of intermediate stages of the DNA–protein complex27,28.
Gene regulation. In a series of incisive experiments, Ptashne and colleagues defined the modus operandi of the lysis/lysogeny decision in phage as a model genetic switch that operates in the right operator sites (oR)29 (Fig. 3). This work has recently been extended by the discovery that the natural switch is strongly influenced by the simultaneous interaction of the repressor with the leftward operator (oL), which is more than 3-kb away30,31,32; this was predicted by the finding that the repressor assembles to form octamers, as required for cooperative binding33.
a | In the bacteriophage life cycle (Fig. 1), the phage can either enter the lytic cyle — in which it reproduces and then lyses its host — or the lysogenic cycle — in which the phage DNA is incorporated into the host DNA and is replicated indefinitely. Dashed boxes enclose operator sites that comprise a promoter-control complex. The three operator sites OR1–3 of the 'λ-switch' control the promoters PRM and PR. Cro and CI dimers bind to the three sites with different affinities and in the opposite order to control the activation of the PRM and PR promoters29. If protein N is available, transcribing RNA polymerases (RNAPs) can be ANTI-TERMINATED at the NUTR and NUTL sites; the termination sites TR1 and TL1 are inoperative for anti-terminated RNAPs. The CI dimer acts as either a repressor or activator of the promoter PRM, depending on its concentration. P1 and P2 are proteases that degrade the CII phage protein. CIII is a phage gene product that is also a substrate for P1. By binding to P1, CIII inhibits the degradation of CII. b | λ-decision-circuit DNA organization. Phage-encoded genetic elements of the decision circuit are located in a 5,000-nucleotide region of the phage DNA. Genes are separated into leftward and rightward transcribed strands as indicated by the arrows. Rightward extensions of the anti-terminated PR transcript transcribe the O and P genes that are essential for phage genome replication, and the Q gene that controls the transcription of later genes on the lytic pathway. Leftward extension of the anti-terminated PL transcript transcribes xis and int genes, which are essential for phage-chromosome integration and excision both into and out of the host chromosome. The locations of four termination sites are indicated by TR1–2 and TL1–2. Figure modified with permission from Ref. 33.
Our initial understanding of in vivo switching was incomplete in another respect. The earlier work showed that the oR switch, by itself, is BISTABLE. The cI gene, which encodes the repressor, is transcribed leftward from oR, whereas the secondary repressor Cro and other gene products come from the rightward message. Both repressor and Cro can bind to three sites in oR and shut-off transcription in both directions. However, one or the other will eventually get the upper hand. If repressor synthesis is established, Cro synthesis is shut off, and vice versa. The system, therefore, provides an elegant model for how switching events might be controlled in other contexts, such as in cell differentiation in multicellular eukaryotes28,34.
As for the phage system, it might seem gratuitous to question whether the crucial events have been identified, as the simple model seems to provide a sufficient explanation. The shut-off of repressor synthesis by Cro depends on Cro binding to the oR3 operator site, which could be the crucial step in activating the lytic cycle. In this view, the establishment of repression depends on a race between Cro and the repressor. However, it has been known from early on that another protein, CII, which acts at the promoter site pRE, is crucial in repressor establishment35 and that Cro that is bound to the other oR sites (oR2 and oR1) decreases CII production. This makes it equally possible that the decisive race is between Cro and CII, with oR3 having, at most, an ancillary role.
The definitive experiments are yet to be done. A comprehensive modelling of the switch is available36, which agrees with the published data on lysogenization frequencies as a function of average phage input37. The complex regulation of the switch is shown in Fig. 3. However, in this pioneering work most of the kinetic parameters are estimates rather than direct measurements. This work underscores the potential for modelling whole-cell physiology with the present high-throughput data on transcription rates and protein abundance. Still lacking is a complete model that predicts the strong overshoot in repressor concentration that is observed during lysogenization34. Because the rate of repressor synthesis from CII-stimulated pRE is much higher than that observed in a lysogen, it seems unlikely that Cro that is bound to oR3 can prevent repressor establishment if CII synthesis achieves its maximum rate.
Tools and pests
Phage therapy. Following their discovery, phages were regarded as possible agents to combat pathogenic bacteria (as discussed by Merril et al.38). Most early attempts at phage therapy were unsuccessful or inconclusive, apparently for trivial technical reasons such as the absence of placebo controls or the rapid clearance of injected phage from the bloodstream before it had time to reach its intended target38,39. With the advent of antibiotics, the development and use of phage therapy almost disappeared from the Western world, although it continued in the Soviet Union and Eastern Europe39. As the incidence of antibiotic resistance continues to rise, the field has been reactivated38. Phage therapy has some inherent advantages over antibiotics, in both the specificity and the ability of the agent to propagate in the desired location (advantages that were seldom realized in earlier work with impure phage preparations)39. As with antibiotics, the efficiency is limited by bacterial mutation, which will doubtless be a great challenge to future development.
At present, phage therapy can be described as promising, but despite its long history its ability to deliver on the promise remains to be fully shown. However, there have been clear successes with experimentally infected animals40,41, and mathematical modelling corroborates the expectations of feasibility42. Much of the earlier work was relatively crude and less controlled, and the whole area was dismissed without a thorough evaluation (largely because of the availability and economy of antibiotic therapy). Now, the numerous potential problems, such as the deleterious effects of the phages themselves, the inactivation of phages by neutralizing antibodies and the genesis of phage-resistant mutants, remain to be rigorously addressed.
Phages as pests in the food industry. In other areas that require bacterial survival under conditions of incomplete sterility, such as industrial fermentation, phages are the enemy. Phage lysis of lactic-acid bacteria has long been a nuisance in cheese factories. Extensive studies on phages that infect lactic-acid bacteria are continuing in many countries43,44,45. A long time 'holy grail' of research that is directed towards the elimination of the problem has been the isolation or engineering of stable bacterial mutants that are resistant to all of the phages that might attack a given culture46,47.
Phage display libraries. There are many occasions on which molecular biologists would like to identify, from a large collection of proteins or peptides, those few that have some special property (such as binding to a low molecular-weight LIGAND or to a specific site on a protein or DNA molecule). It is easy to fractionate the collection on the basis of its binding properties, but if a binding species is rare, it might be impractical to isolate enough material to determine its structure.
In such cases, it is preferable to isolate not the protein or peptide, but rather the DNA that encodes it and can easily be amplified. The simplest way to do this is in a system in which the fractionation process brings the DNA along with the protein it encodes. Surface antigens on bacterial cells can be treated in this way.
A more powerful method is to display the protein or peptide on the surface of a phage. Phage particles are much smaller than bacteria, so larger libraries can be screened. If the library is composed of TRANSLATIONAL FUSIONS to a protein that is abundant on the surface of a phage particle, the sensitivity is high; under conditions of single infection, the binding properties of a phage particle are determined by the gene fusion in its DNA.
Recent examples include fusions of domains of surface antigens of the human pathogen Neisseria meningitidis onto the surface protein of phage T4, for possible use in vaccine construction, and fusions of random five amino-acid sequences to identify possible binding partners for a nuclease that cuts T4 DNA for packaging48,49.
Phages in bacterial pathogenicity
It was discovered early on that in some important bacterial diseases, such as diphtheria50 and botulism51, pathogenicity depends on the presence of certain prophages, which generally encode toxins. Knowledge has accumulated gradually, but the area has recently blossomed for at least two reasons. First, advances in eukaryotic cell biology and bacterial gene regulation allow a more profound understanding of the interaction between pathogenic bacteria and the human cell. Second, whole-genome sequencing of pathogenic bacteria has disclosed the prevalence not only of prophages, but also of pathogenicity islands that are potentially phage-related. So, a framework for incisive studies on phage-mediated diseases has developed52,53.
It is easier to guess the future of the mechanistic studies than that of the evolutionary studies, in which many lines of inquiry converge. Even from the vantage point of whole-genome sequencing, the prevalence of lysogeny might be underestimated because the identification of prophages requires homology to known phages. It has also long been known that complete prophages, which are able to generate plaque-forming particles, frequently deteriorate into incomplete (defective) prophages by changes that range from point mutations to extensive deletions and/or insertions. A particular type of defective phage — specialized transducing phages — can result from faulty excision from the chromosome, which replaces some phage DNA by host DNA adjacent to the insertion site54. This is one mechanism whereby phages might have acquired toxin genes. Another is direct insertion of toxin genes into phage DNA, which might be transposon mediated. The latter possibility is more likely if the toxin gene is not at a terminus of the prophage, but rather in the middle, as in the λ-related coliphage 933W, which encodes a Shiga-like toxin55.
The evolution of bacterial pathogenicity can only be understood by including the role of phages and pathogenicity islands. Prophages and pathogenicity islands seem to be closely related; however, assumptions about the nature of the relationship might be premature. Pathogenicity islands belong to a broader category of DNA known as specialization islands. Specialization islands are blocks of contiguous genes that are distinguished by several criteria. First, they are dedicated to a common function, such as pathogenicity, which is not directly needed for simple survival. Second, they differ from the surrounding DNA in molecular statistics such as the G+C content, codon usage and neighbour relations. Third, they are present in certain strains of a bacterial species and absent from others. Their widespread occurrence has only come to light through whole genome sequencing, and their molecular statistics indicate that such elements have been derived in the relatively recent past from other bacterial species, to which they might be indigenous56. Their intercalation in only certain strains of the species in which they are now observed indicates the operation of an insertion mechanism, such as phage integration or TRANSPOSITION; also, some islands are flanked by phage-insertion sites, which technically qualifies the whole unit, whatever its historical origin, as a defective prophage57,58.
This raises a host of questions for future research. One scenario (perhaps overly facile) is that the whole unit arose in the species of origin by rearrangements of host and phage DNA, that it was introduced into the species in which we now find it by packing into a phage coat, followed by injection and lysogenization, and that, in its present host, it moves from one strain to another by infection and lysogenization. There is, at present, an extreme paucity of evidence to show that the transfer among strains happens by this method rather than by genetic recombination mechanisms that affect all chromosomal genes.
At the other extreme, we might imagine that pathogenicity islands, similar to other genes that are scored as alien59, could move between species by some other mechanism (perhaps TRANSFORMATION or CONJUGATION) and become inserted into the recipient chromosome by transposition or some other mechanism. In this picture, such foreign DNA is frequently found inside prophages or defective prophages simply because, from a bacterial perspective, prophages constitute junk DNA the disruption of which does not affect bacterial survival. Insertion of island DNA into a pre-existing prophage might seem to add an unnecessary step to the mechanism of origin, but that is not the case. The insertion of island DNA into the surrounding phage-related DNA must have happened somewhere sometime, and it frequently now bears no traces of known insertion processes such as transposition.
Further work should illuminate two outstanding questions. First, if this and other 'alien' DNA came from some distantly related donor species of bacteria, what was the donor? The bacterial genomes that have been sequenced so far provide only a handful of examples of lateral transfer in which both donor and recipient might be inferred — perhaps simply because not enough bacteria have yet been sequenced. Second, is the potential mobility of the element in its present host a significant factor in the epidemiology of the diseases?
Reprise
A scientific field derives its interest less from what we know than from the unanswered questions that seem approachable by the available methods. I have tried to indicate some such questions here, many of which have been incubating for years and have been explored through more lines of evidence than could be discussed. However, the time is ripe to seek definitive answers with improved methods. Future research should include the rigorous demonstration of molecular mechanisms, examination of the global impact of phages on the biosphere and studies of the evolution of bacterial hosts.
References
Twort, F. W. An investigation on the nature of ultramicroscopic viruses. Lancet 189, 1241–1243 (1916).
Ellis, E. L. & Delbrück, M. The growth of bacteriophage. J. Gen. Physiol. 22, 365–384 (1939).
Campbell, A. & Botstein, D. in Lambda II (eds Hendrix, R. W., Roberts, J. W., Stahl, F. W. & Weisberg, R. A.) 356–381 (Cold Spring Harbor Laboratory Press, New York, 1983).
Hendrix, R. W. Bacteriophages: evolution of the majority. Theoret. Pop. Biol. 61, 471–480 (2002).
Hendrix, R. W., Smith, M. C., Burns, R. N., Ford, M. E. & Hatfull, G. F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl Acad. Sci. USA 96, 2192–2197 (1999).
Rybchin, V. N. & Svarchevsky, A. N. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol. 33, 895–903 (1999).
Gribaldo, S. & Philippe, H. Ancient phylogenetic relationships. Theoret. Pop. Biol. 61, 391–408 (2002).
Clark, A. J., Inwood, W., Cloutier, T. & Dhillon, T. S. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J. Mol. Biol. 311, 657–679 (2001).
Bamford, D. H., Burnett, R. D. & Stuart, D. J. Evolution of viral structure. Theoret. Pop. Biol. 61, 461–470 (2002).
Wang, L., Junker, D. & Collisson, E. W. Evidence of natural recombination within the S1 gene of infectious bronchitis virus. Viriology 192, 710–716 (1993).
Lai, M. M. C. & Holmes, K. V. in Fundamental Virology (eds Knipe, D. M. & Howley, P. M.) 641–664 (Lippincott, Williams & Wilkins, Philadelphia, 2001).
Wommack, K. E. & Colwell, R. R. Viroplankton: viruses in aquatic ecosystems. Microbial Mol. Biol. Rev. 64, 69–114 (2000).
Wilhelm, S. W. & Suttle, C. A. Viruses and nutrient cycles in the sea. Bioscience 49, 781–787 (1999).
Suttle, C. A. & Chan, A. M. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60, 3167–3174 (1994).
Suttle, C. A. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28, 237–243 (1994).
Fuhrman, J. A. & Suttle, C. A. Viruses in marine planktonic systems. Oceanography 6, 50–62 (1993).
Molineaux, I. J. No syringes please, ejection of DNA from the virion is enzyme driven. Mol. Microbiol. 40, 1–8 (2001).
Zavriev, S. K. & Shemyakin, M. F. RNA polymerase-dependent mechanism for the stepwise T7 phage transport from the virion into E. coli. Nucleic Acids. Res. 10, 1635–1652 (1982).
García, L. R. & Molineux, I. J. Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J. Bacteriol. 177, 4066–4076 (1995).
García, L. R. & Molineux, I. J. Translocation and specific cleavage of bacteriophage T7 DNA in vivo by Eco KI. Proc. Natl Acad. Sci. USA 96, 12430–12435 (1999).
Simpson, A. A. et al. Structure of the bacteriophage φ29 DNA packaging motor. Nature 408, 745–750 (2000).
Smith, D. E. et al. The bacteriophage straight φ29 portal motor can package DNA against a large internal force. Nature 413, 748–752 (2001).
Nunes-Duby, S. E., Azaro, M. A. & Landy, A. Swapping DNA strands and sensing homology without branch migration in λ-site-specific recombination. Curr. Biol. 5, 139–148 (1996).
Nunes-Duby, S. E., Matsumoto, L. & Landy, A. Site-specific recombination intermediates trapped with suicide substrates. Cell 50, 779–788 (1987).
Segall, A. M. & Nash, H. A. Architectural flexibility in λ-site-specific recombination: three alternate conformations channel the attL site into three distinct pathways. Genes Cells 1, 453–463 (1986).
Segall, A. M. Analysis of higher order intermediates and synapsis in the bent-L pathway of bacteriophage λ-site-specific recombination. J. Biol. Chem. 273, 24256–24265 (1998).
Kovach, M. J., Tirumalai, R. & Landy, A. Site-specific photo-cross-linking between λ-integrase and its DNA recombination target. J. Biol. Chem. 277, 14520–14538 (2002).
Wojciak, J. M., Sankar, D., Landy, A. & Chubb, R. T. Arm-site binding for λ-integrase: solution structure and functional characterization of its amino-terminal domain. Proc. Natl Acad. Sci. USA 99, 3434–3439 (2002).
Ptashne, M. A Genetic Switch: Phage and Higher Organisms. 2nd Edn (Blackwell Science, Malden, Massachusetts, and Cell Press, Cambridge, Massachusetts, 1992).
Révet, B., von Wilchen-Bergmann, B., Bessent, H., Barker, A. & Müller-Hill, B. Four dimers of λ-repressor bound to two suitably spaced pairs of λ-operators form octamers and DNA loops over long distances. Curr. Biol. 9, 151–154 (1999).
Dodd, I. B., Perkins, A. J., Tsenitsides, D. & Egan, J. B. Octamerization of λ-cI repressor is needed for effective repression of P(RM) and switching from lysogeny. Genes Dev. 15, 3013–3022 (2001).
Ptashne, M. & Gann, A. Genes and Signals (Cold Spring Harbor Laboratory Press, New York, 2002).
Senear, D. F. et al. The primary self-assembly reaction of bacteriophage cI repressor dimers is to octamer. Biochemistry 32, 6179–6189 (1993).
Darnell, J., Lodish, H. & Baltimore, D. Molecular Cell Biology (Scientific American Books, New York, 1996).
Reichardt, L. & Kaiser, A. D. Control of λ-repressor synthesis. Proc. Natl Acad. Sci. USA 68, 2185–2189 (1971).
Arkin, A., Ross, J. & McAdams, H. H. Stochastic kinetics analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 149, 1633–1648 (1998).
Kourilsky P. Lysogenization by bacteriophage-λ. I. Multiple infection and the lysogenic response. Mol. Gen. Genet. 122, 183–195 (1993).
Merril, C. R. et al. Long-circulating bacteriophage as antibacterial agents. Proc. Natl Acad. Sci. USA 93, 3188–3192 (1996).
Sulalvelidze, A., Alavidze, Z. & Morris, J. G. Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659 (2001).
Smith, H. W. & Huggins, M. B. Successful treatment of experimental E.coli infections in mice using phage is generally superior over antibiotics. J. Gen. Microbiol. 128, 307–318 (1982).
Biswas, B. et al. Bacteriophage therapy rescues mice bacteremic from a vancomycin-resistant Enterococcus faecium. Infect. Immun. 70, 204–210 (2002).
Levin, B. R. & Bull, J. J. Phage therapy revisited: the population biology of a bacterial infection and its treatment with bacteriophages and antibiotics. Am. Nat. 147, 881–898 (1996).
Desiere, F., McShaun, W. M., van Sinderen, D., Ferretti, J. J. & Brussow, H. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic Streptococci: evolutionary implications for prophage–host interactions. Virology 288, 325–341 (2001).
Brussow, H. Phages of dairy bacteria. Ann. Rev. Microbiol. 55, 283–303 (2001).
Barrangow, R., Yoon, S. S., Breidt, F. & Klaenhammer, T. R. Characterization of six Leuconostoc fallax bacteriophages isolated from an industrial sauerkraut fermentation. Appl. Environ. Microbiol. 60, 5452–5458 (2002).
Djordjevic, G. M. & Klaenhammer, T. R. Bacteriophage-triggered defense systems: phage adaptation and design improvements. Appl. Environ. Microbiol. 63, 4370–4376 (1997).
Lucchini, S., Sidotic, J. & Brussow, H. Broad-range bacteriophage resistance in Staphylococcus thermophilus by insertional mutagenesis. Virology 275, 267–277 (2000).
Jiang, J., Abu-Shilbaych, L. & Rao, V. B. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun. 65, 4770 (1997).
Malys, N., Chang, D. Y., Baumann, R. G., Xie, D. & Black, L. W. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late σ-factor (gp55) interaction. J. Mol. Biol. 319, 289–309 (2002).
Freeman, V. J. Studies of the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J. Bacteriol. 61, 675–688 (1951).
Inoue, K. & Iida, H. Conversion of toxigenicity in Clostridium botulinum type C. Jpn. J. Microbial. 14, 87–89 (1970).
Davis, B. M., Kimsey, H. H., Kane, A. V. & Waldor, M. K. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 21, 4240–4249 (2002).
Wagner, P. L. & Waldor, M. K. Bacteriophage control of bacterial virulence. Infection Immun. 70, 3985–3993 (2002).
Campbell, A. Transduction and segregation in Escherichia coli K-12. Virology 4, 366–384 (1957).
Plunkett, G., Rose, D. J., Durfee, T. S. & Blattner, F. D. Sequence of Shiga toxin 2 phage 933W from Escherichia coli 0157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181, 1761–1778 (1998).
Mrázek, J. & Karlin, S. Detecting alien genes in bacterial genomes. Ann. NY Acad. Sci. 870, 314–329 (1999).
Campbell, A. in Mobile DNA II. (eds Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, M.) 1024–1039 (ASM, Washington, 2002).
Hacker, J., Blum-Ohler, G., Muhldorfer, L. & Tschape, H. Pathogenicity islands of virulent bacteria: structure, function and impact on bacterial evolution. Mol. Microbiol. 236, 1088–1097 (1997).
Karlin, S., Campbell, A. M. & Mrázek, J. Comparative DNA analysis across diverse genomes. Ann. Rev. Genet. 32, 185–225 (1998).
Acknowledgements
The author thanks D. Bamford, B. Weisberg and three anonymous reviewers for helpful suggestions.
Author information
Authors and Affiliations
Glossary
- ANALOGUES
-
Genes (or their products) that are not of common ancestry, but which have equivalent functions.
- ANTI-TERMINATION
-
The protein-mediated prevention of the termination of RNA synthesis.
- BISTABLE
-
Having two steady states that are stable to small fluctuations.
- COLIPHAGE
-
A bacteriophage that infects Escherichia coli bacteria.
- CONJUGATION
-
The intercellular transfer of DNA that is mediated by pili, which are surface appendages that are encoded by certain bacterial plasmids and transposons.
- HETERODUPLEX
-
A DNA molecule that is formed by base pairing between strands that are derived from two DNA molecules that are not identical in sequence.
- HOLLIDAY JUNCTION
-
A point at which the strands of two double-stranded DNA molecules exchange partners, which occurs as an intermediate in genetic recombination.
- HOMOLOGUES
-
Genes (or their products) that are descended from a common ancestral gene.
- INTASOME
-
An assemblage of integrase molecules that are bound to their DNA substrate.
- LAMBDOID
-
Belonging to a group of phages that are related to λ.
- LIGAND
-
A molecule that binds non-covalently to another type of molecule.
- LYSOGENIC BACTERIUM
-
A bacterium that harbours phages in a latent form, from which it can be activated to produce infectious phage particles.
- MONOPHYLETIC GROUP
-
A group that contains all the organisms that are descended from a common evolutionary ancestor.
- ORTHOLOGUE
-
The form of a gene in one species that corresponds most directly to a similar gene in another species.
- PROPHAGE
-
The latent form of phage DNA that is present in lysogenic bacteria.
- TEMPERATE PHAGE
-
A phage that is able to form lysogenic bacteria.
- TRANSFORMATION
-
The uptake of exogenous DNA that becomes permanently incorporated into the genome of a cell.
- TRANSLATIONAL FUSION
-
An artificial construct in which the coding regions of two different proteins are juxtaposed so as to generate a single chimeric protein product if translated.
- TRANSPOSITION
-
The transposon-mediated movement of a segment of DNA.
- TYPE 1 RESTRICTION ENZYME
-
A bacterial enzyme that moves along DNA to cleave it far from its specific site of entry.
Rights and permissions
About this article
Cite this article
Campbell, A. The future of bacteriophage biology. Nat Rev Genet 4, 471–477 (2003). https://doi.org/10.1038/nrg1089
Issue Date:
DOI: https://doi.org/10.1038/nrg1089
This article is cited by
-
Gauge your phage: benchmarking of bacteriophage identification tools in metagenomic sequencing data
Microbiome (2023)
-
Application of coliphage as biocontrol agent in combination with gamma irradiation to eliminate multi-drug-resistant E. coli in minimally processed vegetables
Environmental Science and Pollution Research (2023)
-
Resistance of Dickeya solani strain IPO 2222 to lytic bacteriophage ΦD5 results in fitness tradeoffs for the bacterium during infection
Scientific Reports (2022)
-
Bacteriophages and their potential for treatment of gastrointestinal diseases
Nature Reviews Gastroenterology & Hepatology (2022)
-
Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction
The ISME Journal (2017)