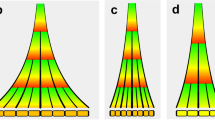
Key Points
-
The leaves of higher plants are derived from the shoot apical meristem.
-
In the Arabidopsis meristem, signalling between CLV and WUS genes creates a feedback loop that maintains the stem-cell population and controls meristem size.
-
Mutually repressive interactions between KNOX proteins and AS1 and AS2 proteins contribute to the delimitation of leaf founder cells in the shoot apex of Arabidopsis.
-
ARP MYB-like proteins are required to repress KNOX expression in both monocot and eudicot plants.
-
Leaf-polarity determinants either promote (adaxial polarity genes PHB/PHV/REV) or antagonize (abaxial polarity genes YAB and KAN) meristem function.
-
Comparative studies of KNOX expression patterns and leaf morphology show a correlation between KNOX expression in leaf primordia and dissected leaf form.
-
KNOX and UFO/LFY define developmental pathways that act in meristems. These might have been co-opted in evolution to function in the generation of the dissected leaf form.
-
Quantitative trait loci analyses indicate that alterations in the regulatory regions of regulatory genes might be responsible for species-specific morphological variation.
Abstract
A key problem in developmental biology is understanding the origin of morphological innovations. Comparative studies in plants with different leaf morphologies indicate that the developmental pathway defined by KNOTTED1-type homeodomain proteins could be involved in generating different leaf forms. The differential expression of regulatory proteins has emerged as an important factor in driving morphological innovations in the plant kingdom — an idea that is well supported by quantitative trait locus analyses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
23 February 2003
genus <i>Fabales</i> corrected to order Fabales
Notes
*A correction was made to this article on the 25 February 2003. The order Fabales was incorrectly referred to as the genus Fabales.
References
Nusslein-Volhard, C. Of flies and fishes. Science 266, 572–574 (1994).
Coen, E. S. & Meyerowitz, E. M. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37 (1991).
Theissen, G. & Saedler, H. The golden decade of molecular floral development (1990–1999): a cheerful obituary. Dev. Genet. 25, 181–193 (1999).
Kramer, E. M. & Irish, V. F. Evolution of genetic mechanisms controlling petal development. Nature 399, 144–148 (1999).
Soltis, D. E. et al. Missing links: the genetic architecture of flower and floral diversification. Trends Plant Sci. 7, 22–31 (2002).
Kenrick, P. & Crane, P. R. The Origin and Early Diversification of Land Plants: A Cladistic Study (Smithsonian Institution, Washington, District of Columbia, 1997).
Theophrastus. Enquiry into Plants (Harvard University Press, Cambridge, Massachusetts and London, UK, 1999).
Bharathan, G. et al. Homologies in leaf form inferred from KNOX1 gene expression during development. Science 296, 1858–1860 (2002). The first comprehensive comparative study of KNOX expression and leaf form, set in a phylogenetic context.
Evans, M. M. S. & Barton, M. K. Genetics of angiosperm shoot apical meristem development. Ann. Rev. Plant Physiol. Plant Mol. Biol. 48, 673–701 (1997).
Clark, S. E. Cell signalling at the shoot meristem. Nature Rev. Mol. Cell Biol. 2, 276–284 (2001).
Fletcher, J. C. Shoot and floral meristem maintenance in Arabidopsis. Annu. Rev. Plant Biol. 53, 45–66 (2002).
Clark, S. E., Running, M. P. & Meyerowitz, E. M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418 (1993).
Clark, S. E., Running, M. P. & Meyerowitz, E. M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067 (1995).
Steeves, T. A. & Sussex, I. M. Patterns in Plant Development (Cambridge University Press, Cambridge, UK, 1989).
Vollbrecht, E., Veit, B., Sinha, N. & Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350, 241–243 (1991).
Vollbrecht, E., Reiser, L. & Hake, S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127, 3161–3172 (2000).
Long, J. A., Moan, E. I., Medford, J. I. & Barton, M. K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 (1996).
Kerstetter, R. et al. Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 6, 1877–1887 (1994).
Smith, L. G., Greene, B., Veit, B. & Hake, S. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116, 21–30 (1992).
Jackson, D., Veit, B. & Hake, S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413 (1994).
Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K. & Hake, S. A Knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876 (1994).
Kerstetter, R. A., Laudencia-Chingcuanco, D., Smith, L. G. & Hake, S. Loss of function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124, 3045–3054 (1997).
Sinha, N. R., Williams, R. E. & Hake, S. Overexpression of the maize homeobox gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7, 787–795 (1993).
Chuck, G., Lincoln, C. & Hake, S. Knat1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289 (1996).
Waites, R., Selvadurai, H. R., Oliver, I. R. & Hudson, A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93, 779–789 (1998).
Timmermans, M. C., Hudson, A., Becraft, P. W. & Nelson, T. ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284, 151–153 (1999).
Tsiantis, M., Schneeberger, R., Golz, J. F., Freeling, M. & Langdale, J. A. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284, 154–156 (1999).
Byrne, M. E. et al. Asymmetric leaves 1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. (2000).
Byrne, M. E., Simorowski, J. & Martienssen, R. A. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965 (2002).
Moore, I., Galweiler, L., Grosskopf, D., Schell, J. & Palme, K. A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl Acad. Sci. USA 95, 376–381 (1998).
Schwabe, W. W. in Positional Controls in Plant Development (eds Barlow, P. W. & Carr, D. J.) 403–440 (Cambridge University Press, Cambridge, UK, 1984).
Reinhardt, D., Mandel, T. & Kuhlemeier, C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507–518 (2000).
Snow, M. & Snow, R. Auxin and leaf formation. New Phytol. 36, 1–18 (1937).
Schwabe, W. W. Chemical modification of phyllotaxis and its implications. Symp. Soc. Exp. Biol. 25, 301–322 (1971).
Galweiler, L. et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230 (1998).
Aida, M., Vernoux, T., Furutani, M., Traas, J. & Tasaka, M. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129, 3965–3974 (2002).
Siegfried, K. R. et al. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128 (1999).
Kerstetter, R. A., Bollman, K., Taylor, R. A., Bomblies, K. & Poethig, R. S. KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709 (2001).
Eshed, Y., Baum, S. F., Perea, J. V. & Bowman, J. L. Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260 (2001).
McConnell, J. R. & Barton, M. K. Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942 (1998).
McConnell, J. R. et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713 (2001).
Talbert, P. B., Adler, H. T., Parks, D. W. & Comai, L. The revoluta gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735 (1995).
Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N. & Clark, S. E. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236 (2001).
Kumaran, M. K., Bowman, J. L. & Sundaresan, V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14, 2761–2770 (2002).
Waites, R. & Hudson, A. phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154 (1995).
Aida, M., Ishida, T. & Tasaka, M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126, 1563–1570 (1999).
Shuai, B., Reynaga-Pena, C. G. & Springer, P. S. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761 (2002).
Elliott, R. C. et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168 (1996).
Long, J. A. & Barton, M. K. The development of apical embryonic pattern in Arabidopsis. Development 125, 3027–3035 (1998). A detailed analysis of early gene-expression patterns during embryogenesis in wild-type Arabidopsis and in stm mutants.
Sentoku, N., Sato, Y. & Matsuoka, M. Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev. Biol. 220, 358–364 (2000).
Coyne, J. A. & Lande, R. The genetic basis of species differences in plants. Am. Nat. 126, 141–145 (1985).
Stark, J., Bonacum, J., Remsen, J. & DeSalle, R. The evolution and development of dipteran wing veins: a systematic approach. Annu. Rev. Entomol. 44, 97–129 (1999).
Hareven, D., Gutfinger, T., Parnis, A., Eshed, Y. & Lifschitz, E. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84, 735–744 (1996). The first demonstration of the relationship between KNOX gene-expression patterns and dissected leaf morphology by expression analysis and KNOX overexpression phenotypes.
Janssen, B. J., Lund, L. & Sinha, N. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117, 771–786 (1998).
Pavlopoulos, A. & Averof, M. Developmental evolution: Hox proteins ring the changes. Curr. Biol. 12, R291–R293 (2002).
Sinha, N. R. Simple and compound leaves: reduction or multiplication? Trends Plant Sci. 2, 396–402 (1997).
Hofer, J., Gourlay, C., Michael, A. & Ellis, T. H. Expression of a class 1 knotted1-like homeobox gene is downregulated in pea compound leaf primordia. Plant Mol. Biol. 45, 387–398 (2001).
Hofer, J. et al. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7, 581–587 (1997). Shows for the first time that UNI — the orthologue of the Arabidopsis LFY protein — regulates leaf form in pea.
Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F. & Meyerowitz, E. M. LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859 (1992).
Coen, E. S. et al. FLORICAULA: a homeotic gene required for flower development in Antirrhinum majus. Cell 63, 1311–1322 (1990).
Taylor, S., Hofer, J. & Murfet, I. I. Stamina pistilloida, the pea ortholog of Fim and UFO, is required for normal development of flowers, inflorescences, and leaves. Plant Cell 13, 31–46 (2001).
Ingram, G. C. et al. Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell 7, 1501–1510 (1995).
Lee, I., Wolfe, D. S., Nilsson, O. & Weigel, D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7, 95–104 (1997).
Molinero-Rosales, N. et al. FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 20, 685–693 (1999).
Estruch, J. J., Prinsen, E., Van Onckelen, H., Schell, J. & Spena, A. Viviparous leaves produced by somatic activation of an inactive cytokinin-synthesizing gene. Science 254, 1364–1367 (1991).
Li, Y., Hagen, G. & Guilfoyle, T. J. Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs. Dev. Biol. 153, 386–395 (1992).
Kusaba, S. et al. Alteration of hormone levels in transgenic tobacco plants overexpressing the rice homeobox gene OSH1. Plant Physiol. 116, 471–476 (1998).
Tanaka-Ueguchi, M., Itoh, H., Oyama, N., Koshioka, M. & Matsuoka, M. Overexpression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J. 15, 391–400 (1998).
Ori, N. et al. Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell 11, 1073–1080 (1999).
Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S. & Matsuoka, M. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15, 581–590 (2001).
Hay, A. et al. The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565 (2002). The authors propose that a regulatory module — defined by KNOX transcription factors and GA — functions in the shoot meristem to control indeterminacy and has been co-opted to function in leaves to control dissected form.
Dover, G. How genomic and developmental dynamics affect evolutionary processes. Bioessays 22, 1153–1159 (2000).
Müller, K. et al. The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 374, 727–730 (1995).
Williams-Carrier, R. E., Lie, Y. S., Hake, S. & Lemaux, P. G. Ectopic expression of the maize kn1 gene phenocopies the Hooded mutant of barley. Development 124, 3737–3745 (1997).
Golz, J. F., Keck, E. J. & Hudson, A. Spontaneous mutations in KNOX genes give rise to a novel floral structure in Antirrhinum. Curr. Biol. 12, 515–522 (2002). The authors characterize two dominant floral mutants caused by regulatory mutations in KNOX genes and propose that similar regulatory changes could be responsible for the evolution of petal spurs in Antirrhinum species.
Muehlbauer, G. J., Fowler, J. E. & Freeling, M. Sectors expressing the homeobox gene liguleless3 implicate a time-dependent mechanism for cell fate acquisition along the proximal–distal axis of the maize leaf. Development 124, 5097–5106 (1997).
Parnis, A. et al. The dominant developmental mutants of tomato, Mouse-Ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell 9, 2143–2158 (1997).
Chen, J. -J., Janssen, B. -J., Williams, A. & Sinha, N. A gene fusion at a homeobox locus: alterations in leaf shape and implications for morphological evolution. Plant Cell 9, 1289–1304 (1997).
Muller, J. et al. In vitro interactions between barley TALE homeodomain proteins suggest a role for protein–protein associations in the regulation of Knox gene function. Plant J. 27, 13–23 (2001).
Bellaoui, M. et al. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13, 2455–2470 (2001).
Smith, H. M., Boschke, I. & Hake, S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl Acad. Sci. USA 99, 9579–9584 (2002).
Burglin, T. R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25, 4173–4180 (1997).
Rieckhof, G. E., Casares, F., Ryoo, H. D., Abu-Shaar, M. & Mann, R. S. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91, 171–183 (1997).
Spit, A., Hyland, R. H., Mellor, E. J. C. & Casselton, L. A. A role for heterodimerization in nuclear localization of a homeodomain protein. Proc. Natl Acad. Sci. USA 95, 6228–6233 (1998).
Lucas, W. J. et al. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270, 1980–1983 (1995).
Kim, J. Y., Yuan, Z., Cilia, M., Khalfan-Jagani, Z. & Jackson, D. Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc. Natl Acad. Sci. USA 99, 4103–4108 (2002).
Perbal, M. -C., Haughn, G., Saedler, H. & Schwarz-Sommer, Z. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development 122, 3433–3441 (1996).
Sessions, A., Weigel, D. & Yanofsky, M. F. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263 (1999).
Helariutta, Y. et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567 (2000).
Ori, N., Eshed, Y., Chuck, G., Bowman, J. L. & Hake, S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532 (2000). Characterization of two mutants that confer ectopic KNOX expression in Arabidopsis leaves and genetic interactions that enhance these mutant phenotypes in a KNOX-independent manner. The results indicate that KNOX expression might be controlled at the chromatin level.
Eshed, Y., Baum, S. F. & Bowman, J. L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209 (1999).
Ogas, J., Kaufmann, S., Henderson, J. & Somerville, C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl Acad. Sci. USA 96, 13839–13844 (1999).
Prigge, M. J. & Wagner, D. R. The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13, 1263–1279 (2001).
Cong, B., Liu, J. & Tanksley, S. D. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl Acad. Sci. USA 99, 13606–13611 (2002). Powerful use of QTL analysis to identify regulatory changes in the fw2.2 locus that are responsible for natural variation in fruit size.
Wang, R. L., Stec, A., Hey, J., Lukens, L. & Doebley, J. The limits of selection during maize domestication. Nature 398, 236–239 (1999). The authors present compelling evidence that selection in regulatory regions of tb1 has driven changes in gene expression and consequent changes in branching patterns during the domestication of maize.
Briggs, D. & Walters, S. M. Plant Variation and Evolution (Cambridge University Press, Cambridge, UK, 1997).
Comai, L. et al. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12, 1551–1568 (2000).
Nasrallah, M. E., Yogeeswaran, K., Snyder, S. & Nasrallah, J. B. Arabidopsis species hybrids in the study of species differences and evolution of amphiploidy in plants. Plant Physiol. 124, 1605–1614 (2000).
Lee, H. S. & Chen, Z. J. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl Acad. Sci. USA 98, 6753–6758 (2001). The analysis of gene-expression profiles during polyploidization reported by this paper and reference 97 indicate that epigenetic regulation might be important in generating diversity during abrupt speciation.
Finnegan, E. J. Epialleles — a source of random variation in times of stress. Curr. Opin. Plant Biol. 5, 101–106 (2002).
Furlong, R. F. & Holland, P. W. Were vertebrates octoploid? Phil. Trans. R. Soc. Lond. B 357, 531–544 (2002).
Cubas, P., Vincent, C. & Coen, E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161 (1999). By showing that an epigenetic mutation in CYC is responsible for differences in floral symmetry in a variant of Linaria , this paper shows the importance of epigenetic regulation in driving changes in form.
Yu, L. P., Simon, E. J., Trotochaud, A. E. & Clark, S. E. POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development 127, 1661–1670 (2000).
Bowman, J. L. et al. The story of CRABS CLAW (or how we learned to love the mutagen). Flowering Newsletter (2001). Excellent exposition of the logic and merits of using second-site mutagenesis to understand shoot development in the post-sequencing era.
Davidson, E. H. et al. A genomic regulatory network for development. Science 295, 1669–1678 (2002).
Brown, C. T. et al. New computational approaches for analysis of cis-regulatory networks. Dev. Biol. 246, 86–102 (2002).
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M. & Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619 (2000).
Schoof, H. et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 (2000).
Riechmann, J. L. et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110 (2000).
Groot, E. P. & Meicenheimer, R. D. Comparison of leaf plastochron index and allometric analyses of tooth development in Arabidopsis thaliana. J. Plant Growth Regul. 19, 77–89 (2000).
Hagemann, W. & Gleissberg, S. Organogenetic capacity of leaves: the significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199, 121–152 (1996). A complete and clearly discussed exposition of the morphological analysis of dissected leaf development in a variety of angiosperms.
Gleissberg, S. in Developmental Genetics and Plant Evolution Vol. 65 (eds Cronk, Q. C., Bateman, R. M. & Hawkins, J. A.) 404–417 (Taylor & Francis, London, UK and New York, 2002).
Kaplan, D. R. Fundamental concepts of leaf morphology and morphogenesis: a contribution to the interpretation of molecular genetic mutants. Int. J. Plant Sci. 162, 465–474 (2001).
Carroll, S., Grenier, J. K. & Weatherbee, S. D. From DNA to Diversity (Blackwell Science, London, UK (2001).
Ronshaugen, M., McGinnis, N. & McGinnis, W. Hox protein mutation and macroevolution of the insect body plan. Nature 415, 914–917 (2002).
Galant, R. & Carroll, S. B. Evolution of a transcriptional repression domain in an insect Hox protein. Nature 415, 910–913 (2002).
Frary, A. et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88 (2000).
Doebley, J., Stec, A. & Hubard, L. The evolution of apical dominance in maize. Nature 386, 485–488 (1997).
Simpson, P. Evolution of development in closely related species of flies and worms. Nature Rev. Genet. 3, 907–917 (2002).
Markstein, M., Markstein, P., Markstein, V. & Levine, M. S. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl Acad. Sci. USA 99, 763–768 (2002).
Fosket, D. E. Plant Growth and Development: a Molecular Approach (Academic Press, San Diego, 1994).
Acknowledgements
We thank N. Sinha for providing figure 1, A Hudson for helpful discussions and S. McCormick for comments on the manuscript. M.T. is a recipient of a Royal Society Fellowship and his laboratory is funded by the Biotechnology and Biological Sciences Research Council.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
TAIR
FURTHER INFORMATION
New York Plant Genomics Consortium
Glossary
- MEGAPHYLLOUS LEAVES
-
The basic leaf type in ferns and seed plants. They are typically large, with a distinct adaxial and abaxial plane and have a complex branched vascular structure.
- MERISTEM
-
A structure that is found at the growing tip of the roots or shoots of a plant and in which stem cells (undifferentiated cells that can undergo unlimited division) are maintained and organogenesis occurs.
- PLASTOCHRON
-
The time interval between successive primordia.
- HIGHER PLANTS
-
Seed plants, including gymnosperms (in which the seed is 'naked') and angiosperms, or flowering plants (in which the seed is contained in an ovary).
- INDETERMINATE
-
Refers to a pattern of growth and development that is open-ended. In meristems, this is associated with the continuous ability to produce determinate lateral organs (such as leaves). By contrast, determinate refers to growth and development that is restricted in time or space.
- LEAF FOUNDER CELLS
-
A group of cells that encompasses several layers of the shoot apical meristem and from which a leaf primordium is derived. Founder cells do not function as stem cells but rather have a determinate fate and can be defined by histological, clonal and molecular criteria.
- LATERAL ORGAN
-
Organs that are produced from the shoot apical meristem such as leaves and putatively homologous organs such as cotyledons (embryonic leaves), bracts (modified leaves that subtend reproductive structures) and floral organs.
- HOMEOBOX
-
A 180-base-pair sequence that encodes a DNA-binding helix–turn–helix motif termed a homeodomain.
- CYCADS
-
(Phylum Cycadophyta). One of four divisions of gymnosperms. They resemble palms but have naked seeds on the scales of cones.
- CLASS I KNOX GENES
-
Knotted1-like homeobox (KNOX) genes in all plants can be divided into two classes based on sequence similarity and expression pattern. The sequenced genome of Arabidopsis contains four class I KNOX genes (STM, KNAT1 (KN1-like in Arabidopsis thaliana 1), KNAT2 and KNAT6).
- MONOCOT
-
(Monocotyledonous plants). Angiosperms with one cotyledon (seed leaf).
- DICOT
-
(Eudicotyledonous plants). The largest group of angiosperms, characterized by two cotyledons (seed leaves) and three symmetrically placed pollen apertures.
- MYB TRANSCRIPTION FACTOR
-
A family of transcription factors defined by the presence of a structurally conserved DNA binding domain first identified in a viral oncogene. The Arabidopsis genome contains over one hundred MYB-like genes that function in diverse regulatory systems, including secondary metabolism, cell morphogenesis, the cell cycle and circadian rhythms.
- HOMEODOMAIN
-
A highly conserved sequence motif. It comprises 60 amino acids and contains a DNA-binding helix–turn–helix motif, indicating that homeodomain-containing proteins function as transcription factors.
- HIGH MOBILITY GROUP (HMG) DOMAIN
-
A conserved domain that is present in HMG proteins, which are non-histone proteins involved in chromatin structure and gene regulation.
- GARP DOMAIN
-
A conserved DNA-binding domain that is distantly related to the MYB DNA-binding domain. It is defined by the acronym of the founding members of the GARP family: maize Golden 2, ARR (Arabidopsis response regulators) and Psr1 (phosphorus stress response1 from Chlamydomonas).
- AUXINS
-
A class of plant hormones, typified by indole-3-acetic acid, that is required for many aspects of plant development and for plant cell growth in culture. The term auxin is derived from the Greek afksano, meaning 'to increase'.
- CAMV 35S PROMOTER
-
35S RNA promoter from cauliflower mosaic virus that is commonly used to drive constitutive gene expression in plants.
- ABAXIAL
-
The side of a lateral organ that initiates away from (ab, away from) the meristem.
- ADAXIAL
-
The side of a lateral organ that initiates next to (ad, close to) the meristem.
- LAMINA
-
The (usually flattened) parts of a leaf on either side of the midvein.
- PINNATE
-
A dissected leaf with a central axis and a single order of leaflets (or pinnae) on either side of it.
- BIPINNATE
-
A dissected leaf that has two orders of leaflets (or pinnae) so the first-order leaflets are themselves dissected into a second order of leaflets.
- PALMATE
-
A dissected leaf with leaflets arising from a central point at the end of the leaf stem
- INTERCALARY LEAFLETS
-
Leaflets that develop between the primary leaflets of a dissected leaf. They are usually smaller than primary leaflets.
- QA REPRESSION DOMAIN
-
A repressive domain that is conserved in insect Ultrabithorax proteins. It contains a Gln-Ala-Gln-Ala-Lys motif and a stretch of Ala residues.
- HETEROCHRONIC REGULATORY MUTATIONS
-
Mutations that alter the relative timing of developmental events as an organism grows (from the Greek heteros, meaning 'other' or 'different', and chronos, meaning 'time').
- AWN
-
A slender bristle-like structure found on the spikelets (structure that contains the flowers) of many grasses.
- PETAL SPUR
-
An outgrowth of a petal in which nectar can collect.
- PLASMODESMATA
-
Cell-wall channels that allow symplastic connections between plant cells.
- NON-AUTONOMOUS
-
Function of a gene product that is not restricted to the cell in which it is expressed.
- PHENOCOPY
-
A mimic of another phenotype.
- CARPEL
-
The female reproductive organ of a flower that encloses the ovules.
Rights and permissions
About this article
Cite this article
Tsiantis, M., Hay, A. Comparative plant development: the time of the leaf?. Nat Rev Genet 4, 169–180 (2003). https://doi.org/10.1038/nrg1002
Issue Date:
DOI: https://doi.org/10.1038/nrg1002
This article is cited by
-
Overexpression of TCP transcription factor OsPCF7 improves agronomic trait in rice
Molecular Breeding (2020)
-
Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum
Molecular Genetics and Genomics (2014)
-
Profiling expression changes caused by a segmental aneuploid in maize
BMC Genomics (2008)
-
Meristem maintenance and compound-leaf patterning utilize common genetic mechanisms in tomato
Planta (2007)
-
Expression of Cell Cycle Genes in Shoot Apical Meristems
Plant Molecular Biology (2006)