Key Points
-
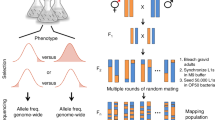
The advantages of the nematode Caenorhabditis elegans for genetics studies are its short generation time, and that it is easy and inexpensive to maintain. Mutant strains can be frozen and maintained indefinitely.
-
Because strains can be propagated as self-fertilizing hermaphrodites, screens for recessive mutations are easy — the F2 generation of mutagenized animals can be inspected for interesting phenotypes. Labour-intensive crosses are not required.
-
Suppressor screens have been widely used to identify components of signal-transduction pathways.
-
The ease with which mutants can be obtained is useful for saturation screening and for the analysis of structure–function relationships of a protein.
-
Some screens are labour intensive, but are nevertheless fruitful if there are no other ways to generate mutants in a specific biological process. These have included screens for cell-death mutants, which have required screening on a compound microscope.
-
Lethal mutations can be identified and maintained in heterozygous animals that segregate 1 in 4 dead embryos. This led to the development of maternal-effect lethal screens, which identified proteins that are required for early cell-fate decisions in the developing embryo.
-
Sensitized screens can be used to identify genes that have a role in a process but are mutated to lethality. The advantage is that the screens can be carried out in the F1 generation and are identified as haploinsufficient loss-of-function loci.
-
Synthetic-lethal screens can identify loci that act redundantly in a process.
Abstract
The nematode Caenorhabditis elegans was chosen as a model genetic organism because its attributes, chiefly its hermaphroditic lifestyle and rapid generation time, make it suitable for the isolation and characterization of genetic mutants. The most important challenge for the geneticist is to design a genetic screen that will identify mutations that specifically disrupt the biological process of interest. Since 1974, when Sydney Brenner published his pioneering genetic screen, researchers have developed increasingly powerful methods for identifying genes and genetic pathways in C. elegans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Ankeny, R. A. The natural history of Caenorhabditis elegans research. Nature Rev. Genet. 2, 474–479 (2001).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).The paper that started it all; it remains a good introduction to C. elegans genetics.
Jin, Y., Hoskins, R. & Horvitz, H. R. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature 372, 780–783 (1994).
Hedgecock, E. M., Culotti, J. G. & Hall, D. H. The unc-5, unc-6, and unc-40 genes guide circumferential migration of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron 2, 61–85 (1990).
Ishii, N., Wadsworth, W. G., Stern, B. D., Culotti, J. G. & Hedgecock, E. M. UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron 9, 873–881 (1992).
Sulston, J. & Horvitz, H. R. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 82, 41–55 (1981).
Ferguson, E. & Horvitz, H. R. Identification and characterization of 22 genes that affect the vulval cell lineages of Caenorhabditis elegans. Genetics 110, 17–72 (1985).A gigantic undertaking that clearly explains the various genetic tests that are used to categorize mutations.
Wang, M. & Sternberg, P. W. Pattern formation during C. elegans vulval induction. Curr. Top. Dev. Biol. 51, 189–220 (2001).
Han, M. & Sternberg, P. W. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell 63, 921–931 (1990).
Beitel, G. J., Clark, S. G. & Horvitz, H. R. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348, 503–509 (1990).
Sternberg, P. W., Golden, A. & Han, M. Role of a raf proto-oncogene during Caenorhabditis elegans vulval development. Phil. Trans. R. Soc. Lond. B Biol. Sci. 340, 259–265 (1993).
Lackner, M. R., Kornfeld, K., Miller, L. M., Horvitz, H. R. & Kim, S. K. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev. 8, 160–173 (1994).
Wu, Y. & Han, M. Suppression of activated LET-60 ras protein defines a role of Caenorhabditis elegans sur-1 MAP kinase in vulval differentiation. Genes Dev. 8, 147–159 (1994).
Kornfeld, K., Hom, D. B. & Horvitz, H. R. The ksr-1 gene encodes a novel protein kinase involved in ras-mediated signaling in C. elegans. Cell 83, 903–913 (1995).
Kornfeld, K., Guan, K.-L. & Horvitz, H. R. The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes Dev. 9, 756–768 (1995).
Singh, N. & Han, M. sur-2, a novel gene, functions late in the let-60 ras-mediated signaling pathway during Caenorhabditis elegans vulval induction. Genes Dev. 9, 2251–2265 (1995).
Sundaram, M. & Han, M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83, 889–901 (1995).
Sternberg, P. W. & Han, M. Genetics of RAS signaling in C. elegans. Trends Genet. 14, 466–472 (1998).A more complete description of the RAS signalling pathway and the history of the genetic screens carried out to identify the genes in this pathway.
Clark, S. G., Stern, M. J. & Horvitz, H. R. Genes involved in two Caenorhabditis elegans cell-signaling pathways. Cold Spring Harb. Symp. Quant. Biol. 57, 363–373 (1992).By suppressing a loss-of-function mutation that constitutively activates the vulval pathway, mutations in the activators of the pathway were obtained.
Hodgkin, J. More sex-determination mutants of Caenorhabditis elegans. Genetics 96, 649–664 (1980).
Miller, L. M., Plenefisch, J. D., Casson, L. P. & Meyer, B. J. xol-1: a gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell 55, 167–183 (1988).
Nusbaum, C. & Meyer, B. J. The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics 122, 579–593 (1989).
Alfonso, A., Grundahl, K., Duerr, J. S., Han, H. P. & Rand, J. B. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 261, 617–619 (1993).
Otsuka, A. J. et al. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron 6, 113–122 (1991).
Hosono, R. et al. The unc-18 gene encodes a novel protein affecting the kinetics of acetylcholine metabolism in the nematode Caenorhabditis elegans. J. Neurochem. 58, 1517–1525 (1992).
Maruyama, I. N. & Brenner, S. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 88, 5729–5733 (1991).
Nonet, M. L., Grundahl, K., Meyer, B. J. & Rand, J. B. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell 73, 1291–1305 (1993).
Nonet, M. L. et al. UNC-11, a Caenorhabditis elegans AP180 homologue, regulates the size and protein composition of synaptic vesicles. Mol. Biol. Cell 10, 2343–2360 (1999).
Harris, T. W., Hartwieg, E., Horvitz, H. R. & Jorgensen, E. M. Mutations in synaptojanin disrupt synaptic vesicle recycling. J. Cell Biol. 150, 589–600 (2000).
Korswagen, H. C., Park, J. H., Ohshima, Y. & Plasterk, R. H. An activating mutation in a Caenorhabditis elegans Gs protein induces neural degeneration. Genes Dev. 11, 1493–1503 (1997).
Berger, A. J., Hart, A. C. & Kaplan, J. M. Gαs-induced neurodegeneration in Caenorhabditis elegans. J. Neurosci. 18, 2871–2880 (1998).
Ambros, V. in C. elegans II (eds Riddle, D. L., Blumenthal, T., Meyer, B. J. & Priess, J. R.) 501–518 (Cold Spring Harbor Laboratory Press, New York, 1997).
Miller, D. M., Niemeyer, C. J. & Chitkara, P. Dominant unc-37 mutations suppress the movement defect of a homeodomain mutation in unc-4, a neural specificity gene in C. elegans. Genetics 135, 741–753 (1993).An elegant screen designed to survey numerous animals and find rare mutations. The screen identified two interacting proteins.
Winnier, A. R. et al. UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes Dev. 13, 2774–2786 (1999).
Klein, R. D. & Meyer, B. J. Independent domains of the SDC-3 protein control sex determination and dosage compensation in C. elegans. Cell 72, 349–364 (1993).
George, S. E., Simokat, K., Hardin, J. & Chisholm, A. D. The VAB-1 Eph receptor tyrosine kinase functions in neural and epithelial morphogenesis in C. elegans. Cell 92, 633–643 (1998).
Chin-Sang, I. D. et al. The ephrin VAB-2/EFN-1 functions in neuronal signaling to regulate epidermal morphogenesis in C. elegans. Cell 99, 781–790 (1999).
Goodman, M. B. et al. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature 415, 1039–1042 (2002).
Ahmed, S. & Hodgkin, J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403, 159–164 (2000).
Hedgecock, E. M., Sulston, J. E. & Thomson, J. N. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220, 1277–1279 (1983).
Ellis, H. M. & Horvitz, H. R. Genetic control of programmed cell death in the nematode C. elegans. Cell 44, 817–829 (1986).
Liu, Q. A. & Hengartner, M. O. The molecular mechanism of programmed cell death in C. elegans. Ann. NY Acad. Sci. 887, 92–104 (1999).
Hengartner, M. O., Ellis, R. E. & Horvitz, H. R. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356, 494–499 (1992).
Hengartner, M. O. & Horvitz, H. R. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 76, 665–676 (1994).
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W. & Prasher, D. C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
Nonet, M. L. Visualization of synaptic specializations in live C. elegans with synaptic vesicle protein–GFP fusions. J. Neurosci. Methods 89, 33–40 (1999).
Schaefer, A. M., Hadwiger, G. D. & Nonet, M. L. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron 26, 345–356 (2000).
Zhen, M., Huang, X., Bamber, B. & Jin, Y. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron 26, 331–343 (2000).
Crump, J. G., Zhen, M., Jin, Y. & Bargmann, C. I. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron 29, 115–129 (2001).
Zallen, J. A., Yi, B. A. & Bargmann, C. I. The conserved immunoglobulin superfamily member SAX-3/Robo directs multiple aspects of axon guidance in C. elegans. Cell 92, 217–227 (1998).
Satterlee, J. S. et al. Specification of thermosensory neuron fate in C. elegans requires ttx-1, a homolog of otd/Otx. Neuron 31, 943–956 (2001).
Hsieh, J. et al. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13, 2958–2970 (1999).
Grant, B. & Hirsh, D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10, 4311–4326 (1999).
Fares, H. & Greenwald, I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nature Genet. 28, 64–68 (2001).
Fares, H. & Greenwald, I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159, 133–145 (2001).
Abrahante, J. E., Miller, E. A. & Rougvie, A. E. Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen::green fluorescent protein fusion gene. Genetics 149, 1335–1351 (1998).
Troemel, E. R., Sagasti, A. & Bargmann, C. I. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387–398 (1999).
Avery, L. & Horvitz, H. R. A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell 51, 1071–1078 (1987).
Avery, L. The genetics of feeding in Caenorhabditis elegans. Genetics 133, 897–917 (1993).
Davis, M. W., Fleischhauer, R., Dent, J. A., Joho, R. H. & Avery, L. A mutation in the C. elegans EXP-2 potassium channel that alters feeding behavior. Science 286, 2501–2504 (1999).
Clark, D. V., Rogalski, R. M., Donati, L. M. & Baillie, D. L. The unc-22(IV) region of Caenorhabditis elegans: genetic analysis of lethal mutations. Genetics 119, 345–353 (1988).
Johnsen, R. C. & Baillie, D. L. Genetic analysis of a major segment [LGV(left)] of the genome of Caenorhabditis elegans. Genetics 129, 735–752 (1991).
Hodgkin, J. What does a worm want with 20,000 genes? Genome Biol. 2, 2008.1–2008.4 (2001).
Herman, R. K. Crossover suppressors and balanced recessive lethals in Caenorhabditis elegans. Genetics 88, 49–65 (1978).
Rosenbluth, R. E. & Baillie, D. L. The genetic analysis of a reciprocal translocation, eT1(III; V), in Caenorhabditis elegans. Genetics 99, 415–428 (1981).
Page, B. D., Zhang, W., Steward, K., Blumenthal, T. & Priess, J. R. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. 11, 1651–1661 (1997).
Schnabel, R. & Priess, J. in C. elegans II (eds Riddle, D. L., Blumenthal, T., Meyer, B. J. & Priess, J. R.) 361–382 (Cold Spring Harbor Laboratory Press, New York, 1997).
Kemphues, K. J. & Strome, S. in C. elegans II (eds Riddle, D. L., Blumenthal, T., Meyer, B. J. & Priess, J. R.) 335–359 (Cold Spring Harbor Laboratory Press, New York, 1997).
Priess, J. R., Schnabel, H. & Schnabel, R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell 51, 601–611 (1987).A successful maternal-effect screen that identified and characterized the first regulator of embryonic cell fate in C. elegans.
Kemphues, K. J., Priess, J. R., Morton, D. G. & Cheng, N. Identification of genes required for cytoplasmic localization of early C. elegans embryos. Cell 52, 311–320 (1988).
Bowerman, B., Eaton, B. A. & Priess, J. R. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68, 1061–1075 (1992).
O'Connell, K. F., Leys, C. M. & White, J. G. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics 149, 1303–1321 (1998).
Ahringer, J. Embryonic tissue differentiation in Caenorhabditis elegans requires dif-1, a gene homologous to mitochondrial solute carriers. EMBO J. 14, 2307–2316 (1995).
Labouesse, M. & Mango, S. E. Patterning the C. elegans embryo: moving beyond the cell lineage. Trends Genet. 15, 307–313 (1999).
Bowerman, B. Embryonic polarity: protein stability in asymmetric cell division. Curr. Biol. 10, R637–R641 (2000).
Gonczy, P. et al. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 144, 927–946 (1999).
Priess, J. R. & Thomson, J. N. Cellular interactions in early C. elegans embryos. Cell 48, 241–250 (1987).
Goutte, C., Hepler, W., Mickey, K. M. & Priess, J. R. aph-2 encodes a novel extracellular protein required for GLP-1-mediated signaling. Development 127, 2481–2492 (2000).
Goutte, C., Tsunozaki, M., Hale, V. A. & Priess, J. R. APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc. Natl Acad. Sci. USA 99, 775–779 (2002).
Simon, M. A., Bowtell, D. D., Dodson, G. S., Laverty, T. R. & Rubin, G. M. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67, 701–716 (1991).
Gu, G., Caldwell, G. A. & Chalfie, M. Genetic interactions affecting touch sensitivity in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 93, 6577–6582 (1996).
Cali, B. M. & Anderson, P. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet. 260, 176–184 (1998).
Yook, K. J., Proulx, S. R. & Jorgensen, E. M. Rules of nonallelic noncomplementation at the synapse in Caenorhabditis elegans. Genetics 158, 209–220 (2001).
Waterston, R. H., Sulston, J. E. & Couson, A. R. in C. elegans II Vol. 33 (eds Riddle, D. L., Blumenthal, T., Meyer, B. J. & Priess, J. R.) 23–45 (Cold Spring Harbor Laboratory Press, New York, 1997).
The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 (1998).
Hodgkin, J. & Herman, R. K. Changing styles in C. elegans genetics. Trends Genet. 14, 352–357 (1998).
Fraser, A. G. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 (2000).
Gonczy, P. et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408, 331–336 (2000).
Thomas, J. H. Thinking about genetic redundancy. Trends Genet. 9, 395–399 (1993).
Zhu, J. et al. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11, 2883–2896 (1997).
Lambie, E. J. & Kimble, J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112, 231–240 (1991).A description of two genes with unique and redundant functions, and how that information was used as the basis for a genetic screen.
Christensen, S., Kodoyianni, V., Bosenberg, M., Friedman, L. & Kimble, J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H). Development 122, 1373–1383 (1996).
Henderson, S. T., Gao, D., Lambie, E. J. & Kimble, J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120, 2913–2924 (1994).
Tax, F. E., Yeargers, J. J. & Thomas, J. H. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 368, 150–154 (1994).
Ferguson, E. & Horvitz, H. R. The multivulva phenotype of certain C. elegans mutants results from defects in two functionally-redundant pathways. Genetics 123, 109–121 (1989).A genetic analysis of an intriguing example of non-homologous redundancy.
Fay, D. S. & Han, M. The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis 26, 279–284 (2000).
Lundquist, E. A. et al. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development 122, 1601–1610 (1996).
Bender, A. & Pringle, J. R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 1295–1305 (1991).
Davies, A. G., Spike, C. A., Shaw, J. E. & Herman, R. K. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics 153, 117–134 (1999).This paper describes an approach that is broadly applicable for isolating mutations in redundant genes.
Fay, D. S., Keenan, S. & Han, M. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16, 503–517 (2002).
De Bono, M. & Bargmann, C. I. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94, 679–689 (1998).
Sulston, J. E. & Horvitz, H. R. Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977).
Sulston, J. E., Schierenberg, E., White, J. G. & Thomson, J. N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983).
Riddle, D. & Albert, P. S. in C. elegans II Vol. 1 (eds Riddle, D. L., Blumenthal, T., Meyer, B. J. & Priess, J. R.) 739–768 (Cold Spring Harbor Laboratory Press, New York, 1997).
Anderson, P. in Caenorhabditis elegans: modern biological analysis of an organism Vol. 48 (eds Epstein, H. F. & Shakes, D. C.) 31–58 (Academic, San Diego, California, 1995).A comprehensive and quantitative discussion of mutagenesis approaches in C. elegans.
Coulson, A., Sulston, J., Brenner, S. & Karn, J. Toward a physical map of the genome of the nematode C. elegans. Proc. Natl Acad. Sci. USA 83, 7821–7825 (1986).
Fire, A. Integrative transformation of Caenorhabditis elegans. EMBO J. 5, 2673–2680 (1986).
Koch, R., Van Luenen, H. G., Van der Horst, M., Thijssen, K. L. & Plasterk, R. H. Single nucleotide polymorphisms in wild isolates of Caenorhabditis elegans. Genome Res. 10, 1690–1696 (2000).
Plasterk, R. H. A. & Van Luenen, H. G. A. M. in C. elegans II Vol. 1 (eds Riddle, D. L., Blumenthal, T., Meyer, B. J. & Priess, J. R.) 97–116 (Cold Spring Harbor Laboratory Press, New York, 1997).
Bessereau, J. L. et al. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413, 70–74 (2001).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Bernstein, E., Denli, A. M. & Hannon, G. J. The rest is silence. RNA 7, 1509–1521 (2001).
Mango, S. E. Stop making nonsense: the C. elegans smg genes. Trends Genet. 17, 646–653 (2001).
Maine, E. M. RNAi as a tool for understanding germline development in Caenorhabditis elegans: uses and cautions. Dev. Biol. 239, 177–189 (2001).
Zwaal, R. R., Broeks, A., Van Meurs, J., Groenen, J. T. & Plasterk, R. H. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl Acad. Sci. USA 90, 7431–7435 (1993).
Jansen, G., Hazendonk, E., Thijssen, K. L. & Plasterk, R. H. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nature Genet. 17, 119–121 (1997).
Liu, L. X. et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 9, 859–867 (1999).
Jansen, G., Hazendonk, E., Thijssen, K. L. & Plasterk, R. H. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nature Genet. 17, 119–121 (1997).
Pujol, N., Bonnerot, C., Ewbank, J. J., Kohara, Y. & Thierry-Mieg, D. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J. Biol. Chem. 276, 11913–11921 (2001).
Acknowledgements
We thank A. Chisholm, R. Herman and M. Labouesse for comments on the manuscript. J. Srinivasan, R. Sommer, E. Troemel, C. Bargmann and L. Kaltenbach provided photographs. P. Anderson, L. Avery, B. Bowerman, S. Clark, M. Han, T. Harris, M. Hengartner, J. Hodgkin, M. Nonet, R. Korswagen, D. Pilgrim, N. Pujol and J. Priess made helpful contributions and suggestions.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
LocusLink
<i>Schizosaccharomyces pombe</i> GeneDB
Wormbase
FURTHER INFORMATION
Encyclopedia of Life Sciences
Caenorhabditis elegans as an experimental organism
Glossary
- GABA NEURON
-
A neuron that releases the inhibitory neurotransmitter GABA (γ-aminobutyric acid).
- FATE MAP
-
The description of the cell divisions from fertilized egg to adult, linked to the eventual anatomical position of the cell in the animal and the differentiated state, or fate, of the cell.
- OPERON
-
A locus consisting of two or more genes that are transcribed as a unit and are expressed in a coordinated manner.
- RNA INTERFERENCE
-
(RNAi). A process by which double-stranded RNA silences specifically the expression of homologous genes through degradation of their cognate mRNA. In worms, a gene can be selectively disabled and its phenotype determined simply by feeding wild-type animals double-stranded RNA.
- ANCHOR CELL
-
A somatic cell in the gonad that induces vulval development in the underlying epidermal cells.
- M4 MOTOR NEURON
-
A motor neuron in the pharynx that is required for the peristaltic movements of the muscle that move food into the grinder.
- BALANCER CHROMOSOME
-
Balancer chromosomes are used in trans to a chromosome that carries a lethal mutation. Such chromosomes carry deleterious mutations, so that heterozygotes have a selective advantage and are easily maintained. They are used as genetic tools because they allow lethal mutations to be propagated indefinitely. In addition, balancer chromosomes frequently contain rearrangements or translocations that disrupt recombination between the homologues.
- mRNA SURVEILLANCE PATHWAY
-
A pathway that recognizes and degrades mRNA molecules that bear nonsense mutations.
- QUANTITATIVE TRAIT
-
A measurable trait that typically depends on the cumulative action of many genes and the environment. [ok?]
Rights and permissions
About this article
Cite this article
Jorgensen, E., Mango, S. The art and design of genetic screens: Caenorhabditis elegans. Nat Rev Genet 3, 356–369 (2002). https://doi.org/10.1038/nrg794
Issue Date:
DOI: https://doi.org/10.1038/nrg794
This article is cited by
-
Screens in fly and beetle reveal vastly divergent gene sets required for developmental processes
BMC Biology (2022)
-
Exogenous misfolded protein oligomers can cross the intestinal barrier and cause a disease phenotype in C. elegans
Scientific Reports (2021)
-
The genome-wide rate and spectrum of EMS-induced heritable mutations in the microcrustacean Daphnia: on the prospect of forward genetics
Heredity (2021)
-
Harnessing the power of genetics: fast forward genetics in Caenorhabditis elegans
Molecular Genetics and Genomics (2021)
-
Analyse von Zellfunktionen mit Hochdurchsatz-Mikroskopie und KI
BIOspektrum (2021)