Key Points
-
Allograft transplantation is a genetically complex medical scenario that requires the consideration of the genomes of two individuals: those of the donor and the recipient.
-
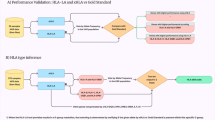
Alloimmunity and histocompatibility are dependent on human leukocyte antigen (HLA) variants, but the role of non-HLA factors has been increasingly recognized in recent years.
-
The majority of genetic studies in the transplantation field have focused on variants in candidate genes, while genome-wide association studies (GWAS) and exome sequencing have just begun to be used in this field.
-
Diagnostic technologies in the transplant field have become increasingly sophisticated alongside the advances in genetic technologies, and they are starting to be used by clinicians.
-
Much larger genetic studies than have previously been conducted will be needed to identify associations with the wide variety of post-transplant outcomes in the many types of organ transplants. Initiatives to collect tissue and data from multiple centres, such as iGeneTRAiN, have been started to address the issue of low sample numbers and non-uniform characterization.
-
The integration of multiple omics technologies will be needed to gain a deeper understanding of the mechanisms and pathways underlying transplant outcomes.
Abstract
Ever since the discovery of the major histocompatibility complex, scientific and clinical understanding in the field of transplantation has been advanced through genetic and genomic studies. Candidate-gene approaches and recent genome-wide association studies (GWAS) have enabled a deeper understanding of the complex interplay of the donor–recipient interactions that lead to transplant tolerance or rejection. Genetic analysis in transplantation, when linked to demographic and clinical outcomes, has the potential to drive personalized medicine by enabling individualized risk stratification and immunosuppression through the identification of variants associated with immune-mediated complications, post-transplant disease or alterations in drug-metabolizing genes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Morris, P. J. Transplantation — a medical miracle of the 20th century. N. Engl. J. Med. 351, 2678–2680 (2004). This is a good perspective article on the medical history of transplantation and the remaining challenges in the field.
Thorsby, E. A short history of HLA. Tissue Antigens 74, 101–116 (2009).
McCaughan, J. A., McKnight, A. J., Courtney, A. E. & Maxwell, A. P. Epigenetics: time to translate into transplantation. Transplantation 94, 1–7 (2012).
Durbin, R. M. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Carja, O., Macisaac, J. L., Mah, S. M., Henn, B. & Kobor, M. S. Worldwide patterns of human genetic and epigenetic variation. Preprint at bioRxiv http://dx.doi.org/10.1101/021931 (2015).
Aubert, O. et al. Long term outcomes of transplantation using kidneys from expanded criteria donors: prospective, population based cohort study. BMJ 351, h3557 (2015).
Klein, J. & Sato, A. The HLA system (first of two parts). N. Engl. J. Med. 343, 702–709 (2000).
Klein, J. & Sato, A. The HLA system (second of two parts). N. Engl. J. Med. 343, 782–786 (2000). References 7 and 8 provide a thorough two-part review of what the HLA system is and how it works.
Phelan, P. J., Conlon, P. J. & Sparks, M. A. Genetic determinants of renal transplant outcome: where do we stand? J. Nephrol. 27, 247–256 (2014).
McVean, G. A. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Schaub, M. A., Boyle, A. P., Kundaje, A., Batzoglou, S. & Snyder, M. Linking disease associations with regulatory information in the human genome. Genome Res. 22, 1748–1759 (2012).
Xie, T. et al. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 13, 2621–2636 (2003).
Flajnik, M. F. & Kasahara, M. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity 15, 351–362 (2001).
Held, P. J. et al. The impact of HLA mismatches on the survival of first cadaveric kidney transplants. N. Engl. J. Med. 331, 765–770 (1994). This is one of the earliest studies showing the importance of matching of specific HLA types for allograft survival.
Wang, C. et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proc. Natl Acad. Sci. USA 109, 8676–8681 (2012).
Ehrenberg, P. K. et al. High-throughput multiplex HLA genotyping by next-generation sequencing using multi-locus individual tagging. BMC Genomics 15, 864 (2014).
Weimer, E. T., Montgomery, M., Petraroia, R., Crawford, J. & Schmitz, J. L. Performance characteristics and validation of next-generation sequencing for human leucocyte antigen typing. J. Mol. Diagn. 18, 668–675 (2016).
Profaizer, T. et al. HLA genotyping in the clinical laboratory: comparison of next-generation sequencing methods. HLA 88, 14–24 (2016).
Effie Petersdorf, W. et al. High HLA-DP expression and graft-versus-host disease. N. Engl. J. Med. 373, 599–609 (2015).
Robinson, J. et al. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43, D423–D431 (2015).
Sigdel, T. K. et al. Non-HLA antibodies to immunogenic epitopes predict the evolution of chronic renal allograft injury. J. Am. Soc. Nephrol. 23, 750–763 (2012).
Terasaki, P. I. Deduction of the fraction of immunologic and non-immunologic failure in cadaver donor transplants. Clin. Transpl. 2003, 449–452 (2003).
Pratschke, J., Weiss, S., Neuhaus, P. & Pascher, A. Review of nonimmunological causes for deteriorated graft function and graft loss after transplantation. Transpl. Int. 21, 512–522 (2008).
Van Bergen, J. et al. KIR-ligand mismatches are associated with reduced long-term graft survival in HLA-compatible kidney transplantation. Am.J. Transplant. 11, 1959–1964 (2011).
Tran, T. H. et al. No impact of KIR-ligand mismatch on allograft outcome in HLA-compatible kidney transplantation. Am. J. Transplant. 13, 1063–1068 (2013).
Mizutani, K. et al. Association of kidney transplant failure and antibodies against MICA. Hum. Immunol. 67, 683–691 (2006).
Zou, Y., Stastny, P., Susal, C., Dohler, B. & Opelz, G. Antibodies against MICA antigens and kidney-transplant rejection. N. Engl. J. Med. 357, 1293–1300 (2007).
Gratwohl, A., Döhler, B., Stern, M. & Opelz, G. H-Y as a minor histocompatibility antigen in kidney transplantation: a retrospective cohort study. Lancet 372, 49–53 (2008).
Kim, S. J. & Gill, J. S. H-Y incompatibility predicts short-term outcomes for kidney transplant recipients. J. Am. Soc. Nephrol. 20, 2025–2033 (2009).
Tan, J. C. et al. H-Y antibody development associates with acute rejection in female patients with male kidney transplants. Transplantation 86, 75–81 (2008).
Yang, J. Y. C., Sigdel, T. K. & Sarwal, M. M. Self-antigens and rejection. Curr. Opin. Organ Transplant. 21, 362–367 (2016).
Porcheray, F. et al. Polyreactive antibodies developing amidst humoral rejection of human kidney grafts bind apoptotic cells and activate complement. Am.J. Transplant. 13, 2590–2600 (2013).
Gao, B. et al. Evidence to support a contribution of polyreactive antibodies to HLA serum reactivity. Transplantation 100, 217–226 (2016).
Subramanian, V. & Mohanakumar, T. Chronic rejection: a significant role for Th17-mediated autoimmune responses to self-antigens. Expert Rev. Clin. Immunol. 8, 663–672 (2012).
Li, L. et al. Identifying compartment-specific non-HLA targets after renal transplantation by integrating transcriptome and 'antibodyome' measures. Proc. Natl Acad. Sci. USA 106, 4148–4153 (2009). This study integrates two different omics technologies to identify organ compartment-specific genes that are targeted by antibodies specific for non-HLA molecules.
Zhang, Q. & Reed, E. F. The importance of non-HLA antibodies in transplantation. Nat. Rev. Nephrol. 12, 484–495 (2016).
Li, L. et al. Compartmental localization and clinical relevance of MICA antibodies after renal transplantation. Transplantation 89, 312–319 (2010).
Sutherland, S. M. et al. Protein microarrays identify antibodies to protein kinase Cζ that are associated with a greater risk of allograft loss in pediatric renal transplant recipients. Kidney Int. 76, 1277–1283 (2009).
Delville, M. et al. A circulating antibody panel for pretransplant prediction of FSGS recurrence after kidney transplantation. Sci. Transl Med. 6, 256ra136 (2014).
Jackson, A. M. et al. Endothelial cell antibodies associated with novel targets and increased rejection. J. Am. Soc. Nephrol. 26, 1161–1171 (2014).
Georgiou, G. et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 32, 158–168 (2014).
Sirota, M., Sigdel, T., Boyd, S., Fire, A. & Sarwal, M. VDJ immune repertoire sequencing predicts patients at risk of alloimmune injury. Am. J. Transplant. 16 (Suppl. 3), abstr. 136 (2016).
Goldfarb-Rumyantzev, A. S. & Naiman, N. Genetic predictors of acute renal transplant rejection. Nephrol. Dial. Transplant. 25, 1039–1047 (2010).
Sankaran, D. et al. Cytokine gene polymorphisms predict acute graft rejection following renal transplantation. Kidney Int. 56, 281–288 (1999).
Singh, R., Srivastava, P., Srivastava, A. & Mittal, R. D. Matrix metalloproteinase (MMP-9 and MMP-2) gene polymorphisms influence allograft survival in renal transplant recipients. Nephrol. Dial. Transplant. 25, 3393–3401 (2010).
Lemos, F. B. et al. The beneficial effects of recipient-derived vascular endothelial growth factor on graft survival after kidney transplantation. Transplantation 79, 1221–1225 (2005).
Ghisdal, L. et al. Genome-wide association study of acute renal graft rejection. Am. J. Transplant. 17, 201–209 (2017). This GWAS is the first conducted in solid-organ transplantation to identify genetic associations with acute rejection.
Sham, P., Bader, J. S., Craig, I., O'Donovan, M. & Owen, M. DNA pooling: a tool for large-scale association studies. Nat. Rev. Genet. 3, 862–871 (2002).
Sanders, M. L. et al. Clinical and genetic factors associated with cutaneous squamous cell carcinoma in kidney and heart transplant recipients. Transplant. Direct 1, e13 (2015).
McCaughan, J. A., McKnight, A. J. & Maxwell, A. P. Genetics of new-onset diabetes after transplantation. J. Am. Soc. Nephrol. 25, 1037–1049 (2014).
Oetting, W. S. et al. Genomewide association study of tacrolimus concentrations in African American kidney transplant recipients identifies multiple CYP3A5 alleles. Am. J. Transplant. 16, 574–582 (2016).
O'Brien, R. P. et al. A genome-wide association study of recipient genotype and medium-term kidney allograft function. Clin. Transplant. 27, 379–387 (2013).
Pihlstrøm, H. K. et al. Single nucleotide polymorphisms and long term clinical outcome in renal transplant patients. A validation study. Am. J. Transplant. 17, 528–533 (2016).
Zhong, H. & Prentice, R. L. Correcting “winner's curse” in odds ratios from genome-wide association findings for major complex human diseases. Genet. Epidemiol. 34, 78–91 (2011).
Sindhi, R. et al. Genetic variants in major histocompatibility complex-linked genes associate with pediatric liver transplant rejection. Gastroenterology 135, 830–839 (2008).
Kawase, T., Nannya, Y., Torikai, H., Yamamoto, G. & Onizuka, M. Identification of human minor histocompatibility antigens based on genetic association with highly parallel genotyping of pooled DNA. Blood 111, 3286–3294 (2008).
Kamei, M. et al. HapMap scanning of novel human minor histocompatibility antigens. Blood 113, 5041–5048 (2009).
Chien, J. W. et al. Improving hematopoietic cell transplant outcomes in a new era of genomic research. Biol. Blood Marrow Transplant. 15, 42–45 (2009).
Hansen, J. A., Chien, J. W., Warren, E. H., Zhao, L. P. & Martin, P. J. Defining genetic risk for graft-versus-host disease and mortality following allogeneic hematopoietic stem cell transplantation. Curr. Opin. Hematol. 17, 483–492 (2010).
Jason Chien, W. et al. Evaluation of published single nucleotide polymorphisms associated with acute GVHD. Blood 119, 5311–5319 (2012).
McCarroll, S. A. et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 40, 1166–1174 (2008).
McCarroll, S. A. et al. Donor–recipient mismatch for common gene deletion polymorphisms in graft-versus-host disease. Nat. Genet. 41, 1341–1344 (2009). This study uses a novel genome-wide array to detect copy number variants, which enabled the identification of novel miHAs in GVHD.
Sato-Otsubo, A. et al. Genome-wide surveillance of mismatched alleles for graft versus host disease in stem cell transplantation. Blood 126, 2752–2764 (2015). This GWAS is the largest conducted in HSCT to date and identified numerous loci that are associated with GVHD.
Ogawa, S. et al. Exploration of the genetic basis of GVHD by genetic association studies. Biol. Blood Marrow Transplant. 15, 39–41 (2009).
Bari, R. et al. Genome-wide single-nucleotide polymorphism analysis revealed SUFU suppression of acute graft-versus-host disease through downregulation of HLA-DR expression in recipient dendritic cells. Sci. Rep. 5, 11098 (2015).
Hong, E. P. & Park, J. W. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 10, 117–122 (2012).
Purcell, S., Cherny, S. S. & Sham, P. C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19, 149–150 (2003).
Genovese, G. et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329, 841–845 (2010).
Olabisi, O. A. et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc. Natl Acad. Sci. USA 113, 830–837 (2016).
Kruzel-Davila, E. et al. APOL1-mediated cell injury involves disruption of conserved trafficking processes. J. Am. Soc. Nephrol. http://dx.doi.org/10.1681/ASN.2016050546 (2016).
Callender, C. O. et al. Blacks as donors for transplantation: suboptimal outcomes overcome by transplantation into other minorities. Transplant. Proc. 40, 995–1000 (2008).
Reeves-Daniel, A. M. et al. The APOL1 gene and allograft survival after kidney transplantation. Am. J. Transplant. 11, 1025–1030 (2011).
Freedman, B. I. et al. Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am. J. Transplant. 15, 1615–1622 (2015).
Freedman, B. I. et al. APOL1 genotype and kidney transplantation outcomes from deceased African American donors. Transplantation 100, 194–202 (2016).
Lee, B. T. et al. The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am. J. Transplant. 12, 1924–1928 (2012).
Grams, M. E. et al. Kidney-failure risk projection for the living kidney-donor candidate. N. Engl. J. Med. 374, 411–421 (2016).
Parsa, A. et al. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 369, 2183–2196 (2013).
Zwang, N. A. et al. APOL1-associated end-stage renal disease in a living kidney transplant donor. Am. J. Transplant. 16, 3568–3572 (2016).
Ojo, A. & Knoll, G. A. APOL1 genotyping of African American deceased organ donors: not just yet. Am. J. Transplant. 15, 1457–1458 (2015).
Ross, L. F. & Thistlethwaite, J. R. Introducing genetic tests with uncertain implications in living donor kidney transplantation: ApoL1 as a case study. Prog. Transplant. 26, 203–206 (2016).
Freedman, B. I. & Julian, B. A. Should kidney donors be genotyped for APOL1 risk alleles? Kidney Int. 87, 671–673 (2015).
Cohen, D. M., Mittalhenkle, A., Scott, D. L., Young, C. J. & Norman, D. J. African American living-kidney donors should be screened for APOL1 risk alleles. Transplantation 92, 722–725 (2011).
Chandraker, A. The real world impact of APOL1 variants on kidney transplantation. Transplantation 100, 16–17 (2016).
Ma, J. et al. Deceased donor multidrug resistance protein 1 and caveolin 1 gene variants may influence allograft survival in kidney transplantation. Kidney Int. 88, 584–592 (2015).
Oetting, W. S. et al. Donor polymorphisms of Toll-like receptor 4 associated with graft failure in liver transplant recipients. Liver Transplant. 18, 1399–1405 (2012).
Jacobson, P. A. et al. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation 91, 300–308 (2011).
Tavira, B. et al. Mitochondrial DNA haplogroups and risk of new-onset diabetes among tacrolimus-treated renal transplanted patients. Gene 538, 195–198 (2014).
Farh, K. K.-H. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
[No authors listed]. On beyond GWAS. Nat. Genet. 42, 551–551 (2010).
Thomson, R. et al. Evolution of the primate trypanolytic factor APOL1. Proc. Natl Acad. Sci. USA 111, E2130–E2139 (2014).
Dorr, C. R. et al. Deceased-donor apolipoprotein L1 renal-risk variants have minimal effects on liver transplant outcomes. PLoS ONE 11, e0152775 (2016).
Oetting, W. S. et al. Validation of single nucleotide polymorphisms associated with acute rejection in kidney transplant recipients using a large multi-center cohort. Transpl. Int. 24, 1231–1238 (2011).
Cole, B., van Setten, J. & Keating, B. The distance between us: the landscape of recipient-specific loss-of-function in solid organ transplantation and association with rejection-free graft survival. Am. J. Transplant. 16 (Suppl. 3), abstr. D304 (2016).
Reindl-Schwaighofer, R. et al. Alloimmunity through non-HLA epitopes in kidney transplantation. Am.J. Transplant. 16 (Suppl. 3), abstr. D30 (2016).
Köttgen, A. et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 41, 712–717 (2009).
Menon, M. C. et al. Intronic locus determines SHROOM3 expression and potentiates renal allograft fibrosis. J. Clin. Invest. 125, 208–221 (2015).
Sampson, J. K. et al. Whole exome sequencing to estimate alloreactivity potential between donors and recipients in stem cell transplantation. Br. J. Haematol. 166, 566–570 (2014).
Mesnard, L. et al. Exome sequencing and prediction of long-term kidney allograft function. PLoS Comput. Biol. 12, e1005088 (2015).
Todd, J. L. et al. Whole exome sequencing: a novel strategy to understand chronic lung allograft dysfunction (CLAD). J. Heart Lung Transplant. 33, S140 (2014).
Shreders, A., Asmann, Y., Wang, X. & Roy, V. Using whole exome sequencing to identify genetic variation and polymorphisms associated with graft versus host disease in allogeneic stem cell transplant recipients. Blood 126, 5414 (2015).
Keating, B., Li, Y., Olthoff, K., Wang, J. & Shaked, A. Application of second generation sequencing and AlloAntibody screening to the organ transplantation arena. J. Am. Transplant. 15 (Suppl. 3), abstr. 436 (2015).
Dorr, C. et al. Differentially expressed gene transcripts using RNA sequencing from the blood of immunosuppressed kidney allograft recipients. PLoS ONE 10, e0125045 (2015).
Lin, Y. et al. RNA-seq analysis of clinical-grade osteochondral allografts reveals activation of early response genes. J. Orthop. Res. 34, 1950–1959 (2016).
Gregson, A. L. et al. Altered exosomal RNA profiles in bronchoalveolar lavage from lung transplants with acute rejection. Am. J. Respir. Crit. Care Med. 192, 1490–1503 (2015).
Ben-Dov, I. et al. MicroRNA sequence profiles of human kidney allograft with or without tubulointerstitial fibrosis. Transplantation 94, 1086–1094 (2012).
Naesens, M., Kuypers, D. R. J. & Sarwal, M. Calcineurin inhibitor nephrotoxicity. Clin. J. Am. Soc. Nephrol. 4, 481–508 (2009).
Staatz, C., Taylor, P. & Tett, S. Low tacrolimus concentrations and increased risk of early acute rejection in adult renal transplantation. Nephrol. Dial. Transplant. 16, 1905–1909 (2001).
Relling, M. V. & Evans, W. E. Pharmacogenomics in the clinic. Nature 526, 343–350 (2015).
Hewett, M. et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 30, 163–165 (2002).
Zhang, X. et al. Influence of CYP3A5 and MDR1 polymorphisms on tacrolimus concentration in the early stage after renal transplantation. Clin. Transplant. 19, 638–643 (2005).
Vafadari, R. et al. Genetic polymorphisms in ABCB1 influence the pharmacodynamics of tacrolimus. Ther. Drug Monit. 35, 459–465 (2013).
Hustert, E. et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 11, 773–779 (2001).
Kuehl, P. et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 27, 383–391 (2001).
Pallet, N. et al. Kidney transplant recipients carrying the CYP3A4*22 allelic variant have reduced tacrolimus clearance and often reach supratherapeutic tacrolimus concentrations. Am. J. Transplant. 15, 800–805 (2015).
Lamba, J., Hebert, J. M., Schuetz, E. G., Klein, T. E. & Altman, R. B. PharmGKB summary: very important pharmacogene information for CYP3A5. Pharmacogenet. Genomics 22, 555–558 (2012).
Birdwell, K. A. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin. Pharmacol. Ther. 98, 19–24 (2015). These guidelines represent the integration of knowledge from numerous studies on CYP3A5 genotype and aim to change clinical practice in transplantation.
Passey, C. et al. Validation of tacrolimus equation to predict troughs using genetic and clinical factors. Pharmacogenomics 13, 1141–1147 (2012).
Sanghavi, K. et al. Genotype-guided tacrolimus dosing in African-American kidney transplant recipients. Pharmacogenomics J. 17, 61–68 (2015).
Pallet, N. et al. Long-term clinical impact of adaptation of initial tacrolimus dosing to CYP3A5 genotype. Am. J. Transplant. 16, 2670–2675 (2016).
Rojas, L. et al. Effect of CYP3A5*3 on kidney transplant recipients treated with tacrolimus: a systematic review and meta-analysis of observational studies. Pharmacogenomics J. 15, 38–48 (2015).
Macher, H. C. et al. Monitoring of transplanted liver health by quantification of organ-specific genomic marker in circulating DNA from receptor. PLoS ONE 9, e113987 (2014).
Moreira, V. G., García, B. P., Martín, J. M. B., Suárez, F. O. & Alvarez, F. V. Cell-free DNA as a noninvasive acute rejection marker in renal transplantation. Clin. Chem. 55, 1958–1966 (2009).
Sigdel, T. K. et al. A rapid noninvasive assay for the detection of renal transplant injury. Transplantation 96, 97–101 (2013).
Josephson, M. A. Monitoring and managing graft health in the kidney transplant recipient. Clin. J. Am. Soc. Nephrol. 6, 1774–1780 (2011).
Lo, D. J., Kaplan, B. & Kirk, A. D. Biomarkers for kidney transplant rejection. Nat. Rev. Nephrol. 10, 215–225 (2014).
Viklicky, O., Hribova, P. & Brabcova, I. Molecular markers of rejection and tolerance: lessons from clinical research. Nephrol. Dial. Transplant. 28, 2701–2708 (2013).
Suthanthiran, M. et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N. Engl. J. Med. 190, 2175–2176 (2013).
Keslar, K. S. et al. Multicenter evaluation of a standardized protocol for noninvasive gene expression profiling. Am. J. Transplant. 13, 1891–1897 (2013).
Roedder, S. et al. The kSORT assay to detect renal transplant patients at high risk for acute rejection: results of the multicenter AART study. PLoS Med. 11, e1001759 (2014).
Gimino, V. J., Lande, J. D., Berryman, T. R., King, R. A. & Hertz, M. I. Gene expression profiling of bronchoalveolar lavage cells in acute lung rejection. Am. J. Respir. Crit. Care Med. 168, 1237–1242 (2003).
Morgun, A. et al. Molecular profiling improves diagnoses of rejection and infection in transplanted organs. Circ. Res. 98, e74–e83 (2006).
Halloran, P. F. et al. Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: an international prospective study (INTERCOM). Am. J. Transplant. 13, 2865–2874 (2013).
Sellarés, J. et al. Molecular diagnosis of antibody-mediated rejection in human kidney transplants. Am. J. Transplant. 13, 971–983 (2013).
Halloran, P. F., Merino Lopez, M. & Barreto Pereira, A. Identifying subphenotypes of antibody-mediated rejection in kidney transplants. Am. J. Transplant. 16, 908–920 (2016).
Li, L. et al. A peripheral blood diagnostic test for acute rejection in renal transplantation. Am. J. Transplant. 12, 2710–2718 (2012).
Li, L. et al. Identification of common blood gene signatures for the diagnosis of renal and cardiac acute allograft rejection. PLoS ONE 8, e82153 (2013).
De Vlaminck, I. et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl Med. 6, 241ra77 (2014). This is the first study of its kind in transplantation, and it validates the use of cfDNA as a marker of organ transplant rejection.
Snyder, T. M., Khush, K. K., Valantine, H. A. & Quake, S. R. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl Acad. Sci. USA 108, 6229–6234 (2011).
De Vlaminck, I. et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl Acad. Sci. USA 112, 13336–13341 (2015).
Beck, J. et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin. Chem. 59, 1732–1741 (2013).
Aljurf, M. et al. Chimerism analysis of cell-free DNA in patients treated with hematopoietic stem cell transplantation may predict early relapse in patients with hematologic malignancies. Biotechnol. Res. Int. 2016, 8589270 (2016).
Burnham, P. et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci. Rep. 6, 27859 (2016).
Burra, P. & De Bona, M. Quality of life following organ transplantation. Transpl. Int. 20, 397–409 (2007).
Xiong, J. et al. Lack of association between interleukin-10 gene polymorphisms and graft rejection risk in kidney transplantation recipients: a meta-analysis. PLoS ONE 10, e0127540 (2015).
Liu, F. et al. Interleukin-10-1082G/A polymorphism and acute liver graft rejection: a meta-analysis. World J. Gastroenterol. 18, 847–854 (2012).
Arora, M. et al. Validation study failed to confirm an association between genetic variants in the base excision repair pathway and transplant-related mortality and relapse after hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 22, 1531–1532 (2016).
Thyagarajan, B. et al. Association between genetic variants in the base excision repair pathway and outcomes after hematopoietic cell transplantations. Biol. Blood Marrow Transplant. 16, 1084–1089 (2010).
International Genetics & Translational Research in Transplantation Network. Design and implementation of the International Genetics and Translational Research in Transplantation Network. Transplantation 99, 2401–2412 (2015). This paper reports on the newly conceived iGeneTRAiN initiative, which seeks to alleviate many of the sample size and reproducibility issues found in transplantation studies by establishing an international consortium.
Li, Y. R. et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med. 7, 90 (2015). This paper delineates the creation of a transplantation-specific GWAS array that will be used on samples collected by the iGeneTRAiN initiative.
Sigdel, T. K. et al. Mining the human urine proteome for monitoring renal transplant injury. Kidney Int. 89, 1244–1252 (2016).
Crespo, E. et al. Molecular and functional noninvasive immune monitoring in the ESCAPE study for prediction of subclinical renal allograft rejection. Transplantation http://dx.doi.org/10.1097/TP.0000000000001287 (2016). This paper reports a study of one of the earliest clinically available gene expression-based non-invasive diagnostics in the field of kidney transplantation.
Grskovic, M. et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J. Mol. Diagn. 18, 890–902 (2016).
Pham, M. X. et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N. Engl. J. Med. 362, 1890–1900 (2010).
Sarwal, M. M. et al. Transplantomics and biomarkers in organ transplantation: a report from the first international conference. Transplantation 91, 379–382 (2011).
Nakorchevsky, A. et al. Molecular mechanisms of chronic kidney transplant rejection via large-scale proteogenomic analysis of tissue biopsies. J. Am. Soc. Nephrol. 21, 362–373 (2010).
Akhtar, M. Z. et al. Using an integrated -omics approach to identify key cellular processes that are disturbed in the kidney after brain death. Am. J. Transplant. 16, 1421–1440 (2016).
Gehlenborg, N. et al. Visualization of omics data for systems biology. Nat. Methods 7, S56–S68 (2010).
Lee, D. et al. Rapid determination of Perv copy number from porcine genomic DNA by real-time polymerase chain reaction. Anim. Biotechnol. 22, 175–180 (2011).
Oriol, R., Ye, Y., Koren, E. & Cooper, D. K. Carbohydrate antigens of pig tissues reacting with human natural antibodies as potential targets for hyperacute vascular rejection in pig-to-man organ xenotransplantation. Transplantation 56, 1433–1442 (1993).
Yang, L. et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 350, 1101–1104 (2015).
Sato, M. et al. The combinational use of CRISPR/Cas9-based gene editing and targeted toxin technology enables efficient biallelic knockout of the α-1,3-galactosyltransferase gene in porcine embryonic fibroblasts. Xenotransplantation 21, 291–300 (2014).
Billingham, R. E., Brent, L. & Medawar, P. B. 'Actively acquired tolerance' of foreign cells. Nature 172, 603–606 (1953). This study reports the discovery of acquired tolerance, work for which the authors received the 1953 Nobel Prize.
Wood, K. K. & Sakaguchi, S. Regulatory T cells in transplantation tolerance. Nat. Rev. Immunol. 3, 200–210 (2003).
Luan, Y. et al. Monocytic myeloid-derived suppressor cells accumulate in renal transplant patients and mediate CD4+Foxp3+ Treg expansion. Am. J. Transplant. 13, 3123–3131 (2013).
Drujont, L. et al. Evaluation of the therapeutic potential of bone marrow-derived myeloid suppressor cell (MDSC) adoptive transfer in mouse models of autoimmunity and allograft rejection. PLoS ONE 9, e100013 (2014).
Tang, Q. & Bluestone, J. A. Regulatory T-cell therapy in transplantation: moving to the clinic. Cold Spring Harb. Perspect. Med. 3, a015552 (2013).
Brouard, S. et al. Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance. Proc. Natl Acad. Sci. USA 104, 15448–15453 (2007).
Chesneau, M. et al. Tolerant kidney transplant patients produce B cells with regulatory properties. J. Am. Soc. Nephrol. 26, 2588–2598 (2015).
Roedder, S. et al. A three-gene assay for monitoring immune quiescence in kidney transplantation. J. Am. Soc. Nephrol. 26, 2042–2053 (2014).
Kawai, T. et al. Long-term results in recipients of combined HLA-mismatched kidney and bone marrow transplantation without maintenance immunosuppression. Am. J. Transplant. 14, 1599–1611 (2014).
Leventhal, J. R. et al. Immune reconstitution/immunocompetence in recipients of kidney plus hematopoietic stem/facilitating cell transplants. Transplantation 99, 288–298 (2015).
Le Guen, V. et al. Alloantigen gene transfer to hepatocytes promotes tolerance to pancreatic islet graft by inducing CD8+ regulatory T cells. J. Hepatol. http://dx.doi.org/10.1016/j.jhep.2016.11.019 (2016).
Jindra, P. T., Tripathi, S., Tian, C., Iacomini, J. & Bagley, J. Tolerance to MHC class II disparate allografts through genetic modification of bone marrow. Gene Ther. 20, 478–486 (2012).
Morris, H. et al. Tracking donor-reactive T cells: evidence for clonal deletion in tolerant kidney transplant patients. Sci. Transl Med. 7, 272ra10 (2015).
Rebollo-Mesa, I. et al. Biomarkers of tolerance in kidney transplantation: are we predicting tolerance or response to immunosuppressive treatment? Am. J. Transplant. 16, 3443–3457 (2016).
Sagoo, P. et al. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J. Clin. Invest. 120, 1848–1861 (2010).
Roedder, S., Gao, X. & Sarwal, M. M. The pits and pearls in translating operational tolerance biomarkers into clinical practice. Curr. Opin. Organ Transplant. 17, 655–662 (2012).
Martínez-Llordella, M. et al. Using transcriptional profiling to develop a diagnostic test of operational tolerance in liver transplant recipients. J. Clin. Invest. 118, 2845–2857 (2008).
Bohne, F. et al. Intra-graft expression of genes involved in iron homeostasis predicts the development of operational tolerance in human liver transplantation. J. Clin. Invest. 122, 368–382 (2012).
Anglicheau, D. et al. MicroRNA expression profiles predictive of human renal allograft status. Proc. Natl Acad. Sci. USA 106, 5330–5335 (2009).
Sui, W. et al. Microarray analysis of microRNA expression in acute rejection after renal transplantation. Transpl. Immunol. 19, 81–85 (2008).
Danger, R. et al. Expression of miR-142-5p in peripheral blood mononuclear cells from renal transplant patients with chronic antibody-mediated rejection. PLoS ONE 8, e60702 (2013).
Matz, M. et al. Free microRNA levels in plasma distinguish T-cell mediated rejection from stable graft function after kidney transplantation. Transpl. Immunol. 39, 52–59 (2016).
Oghumu, S. et al. Acute pyelonephritis in renal allografts: a new role for microRNAs? Transplantation 97, 559–568 (2014).
Shaked, A. et al. An ectopically expressed serum miRNA signature is prognostic, diagnostic, and biologically related to liver allograft rejection. Hepatology 65, 269–280 (2017).
Xu, Z., Sharma, M., Gelman, A., Hachem, R. & Mohanakumar, T. Significant role for microRNA-21 affecting Toll-like receptor pathway in primary graft dysfunction after human lung transplantation. J. Heart Lung Transplant. 36, 331–339 (2016).
Van Huyen, J. P. D. et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur. Heart J. 35, 3194–3202 (2014).
Asaoka, T. et al. MicroRNA signature of intestinal acute cellular rejection in formalin-fixed paraffin-embedded mucosal biopsies. Am. J. Transplant. 12, 458–468 (2012).
Kanak, M. A. et al. Evaluation of microRNA375 as a novel biomarker for graft damage in clinical islet transplantation. Transplantation 99, 1568–1573 (2015).
Ulbing, M. et al. MicroRNAs 223-3p and 93-5p in patients with chronic kidney disease before and after renal transplantation. Bone 95, 115–123 (2017).
Gunasekaran, M. et al. Donor-derived exosomes with lung self-antigens in human lung allograft rejection. Am. J. Transplant. 17, 474–484 (2017).
Scian, M. J. et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am. J. Transplant. 11, 2110–2122 (2011).
Wei, L., Gong, X., Martinez, O. M. & Krams, S. M. Differential expression and functions of microRNAs in liver transplantation and potential use as non-invasive biomarkers. Transpl. Immunol. 29, 123–129 (2013).
Glowacki, F. et al. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS ONE 8, e58014 (2013).
Chau, B. N. et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl Med. 4, 121ra18 (2012).
Wilflingseder, J. et al. MiRNA profiling discriminates types of rejection and injury in human renal allografts. Transplantation 95, 835–841 (2013).
Gupta, S. K. et al. MiR-21 promotes fibrosis in an acute cardiac allograft transplantation model. Cardiovasc. Res. 110, 215–226 (2016).
Huibers, M. M. H. et al. Changes of plasma microRNAs in heart transplantation patients do not reflect microRNA changes in the cardiac allograft vasculopathy vessel wall. J. Heart Lung Transplant. 32, S242 (2013).
Su, S. et al. MiR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat. Commun. 6, 8523 (2015).
Godwin, J. G. et al. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc. Natl Acad. Sci. USA 107, 14339–14344 (2010).
Day, E., Kearns, P. K., Taylor, C. J. & Bradley, J. A. Transplantation between monozygotic twins. Transplantation 98, 485–489 (2014).
Rodriguez, R. M. et al. DNA methylation dynamics in blood after hematopoietic cell transplant. PLoS ONE 8, e56931 (2013).
Paul, D. S. et al. A donor-specific epigenetic classifier for acute graft-versus-host disease severity in hematopoietic stem cell transplantation. Genome Med. 7, 128 (2015).
Mehta, T. K. et al. Quantitative detection of promoter hypermethylation as a biomarker of acute kidney injury during transplantation. Transplant. Proc. 38, 3420–3426 (2006).
Lehmann-Werman, R. et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl Acad. Sci. USA 113, E1826–E1834 (2016).
Sun, K. et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl Acad. Sci. USA 112, 201508736 (2015). This study delineates a method that can identify the organ-specific origin of circulating cfDNA in transplantation.
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT02088931 (2016).
Hoffmann, P. et al. Loss of FOXP3 expression in natural human CD4+CD25+ regulatory T cells upon repetitive in vitro stimulation. Eur. J. Immunol. 39, 1088–1097 (2009).
Tao, R. et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13, 1299–1307 (2007).
Van Loosdregt, J. et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood 115, 965–974 (2010).
Benichou, G. Direct and indirect antigen recognition: the pathways to allograft immune rejection. Front. Biosci. 4, D476–D480 (1999).
Benichou, G. Direct versus indirect allorecognition pathways: on the right track. Am. J. Transplant. 9, 655–656 (2009).
Conlon, T. M. et al. Germinal center alloantibody responses are mediated exclusively by indirect-pathway CD4 T follicular helper cells. J. Immunol. 188, 2643–2652 (2012).
Sayegh, M. H., Khoury, S. J., Hancock, W. W., Weiner, H. L. & Carpenter, C. B. Induction of immunity and oral tolerance to alloantigen by polymorphic class II major histocompatibility complex allopeptides in the rat. Transplant. Proc. 25, 357–358 (1993).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
M.M.S. is the founder of ORGAN-I (California, USA), a spin-out company from Stanford University (California, USA) that developed the kSORT assay for the assessment of acute kidney transplant rejection using a blood test. M.M.S. is a scientific advisory board member for Immucor (Georgia, USA), a diagnostic company that is commercializing the kSORT assay.
Related links
FURTHER INFORMATION
Glossary
- Tolerance
-
A state of immune unresponsiveness and quiescence towards specific antigens. In the case of transplantation, tolerance is directed towards donor-specific antigens.
- Allogeneic
-
A term that describes tissues that are of distinct genetic origins and thus often immunologically incompatible.
- Acute rejection
-
An episode of sudden deterioration in allograft function as a result of either antibody-mediated rejection or T cell-mediated rejection, which result from different molecular processes.
- Isograft
-
A graft between two individuals who are genetically identical, such as in the case of monozygotic twins.
- Allografts
-
Grafts from another member of the same species, such as in the case of organ transplantation, as opposed to grafts from a member of a different species (xenograft) or from the recipient themselves (autograft).
- Ischaemic and reperfusion damage
-
Damage to an organ as a result of a transient inadequate blood supply.
- Delayed graft function
-
A state in which renal failure persists after transplantation, thus necessitating dialysis.
- New-onset diabetes after transplantation
-
(NODAT). The occurrence of diabetes mellitus after transplantation in a patient who did not have the disease before. This occurs in 2–53% of all solid-organ transplants and is due in part to the immunosuppressive medications that are used to prevent transplant rejection.
- Alloimmunity
-
An immune response to antigens that are both non-self and from the same species.
- Allorecognition
-
The ability of a host to recognize allogeneic tissue as distinct from its own.
- Complement system
-
A component of the innate immune system that can be activated by antigen-bound antibodies.
- Central tolerance
-
The mechanisms by which T cells and B cells are rendered non-reactive to an antigen, typically a self-antigen, in the primary lymphoid organs.
- Peripheral tolerance
-
The mechanisms by which T cells and B cells are rendered non-reactive to an antigen outside the primary lymphoid organs.
- Regulatory T cells
-
(Treg cells). A subpopulation of T cells that are generally immunosuppressive rather than pro-inflammatory.
- Tolerogenic
-
The quality of being able to induce immunological tolerance.
- Survival
-
In the context of this Review, an outcome measured as the time until either graft failure (when referring to allograft survival) or patient mortality (when referring to recipient survival).
- Ambiguity rates
-
The rates at which sequenced regions have a low level of confidence in excluding possible allelic variations.
- Whole-exome sequencing
-
Sequencing of all protein- coding regions in the genome.
- Whole-genome sequencing
-
Sequencing of the complete DNA genetic material in a cell or organism.
- Graft-versus-host disease
-
(GVHD). Largely specific to haematopoietic stem cell transplantation, it is a medical complication in which immune cells in the donated tissue reject and attack the host cells.
- Killer-cell immunoglobulin-like receptors
-
(KIRs). Receptors that are expressed on the surface of natural killer cells and modulate their cytotoxic activity by recognizing major histocompatibility complex class I allelic variants.
- MHC class I polypeptide-related sequence A
-
(MICA). A cell-surface antigen that is recognized by the receptor NKG2D, which is found on natural killer cells, T cells and macrophages.
- Minor histocompatibility antigens
-
(miHAs). The distinct peptide products of polymorphic genes that distinguish the recipient from the donor.
- Cryptic antigens
-
Self-antigens that are not clonally deleted in the thymus owing to low surface presentation on antigen-presenting cells (APCs). These self-antigens can be expressed by APCs following differential processing by inflammatory proteases.
- Trough levels
-
The lowest levels of a pharmaceutical present in the blood before the next dose.
- Long-term graft function
-
Refers to the functional characteristics of the transplanted organ, which typically decrease over time owing to immune injury and subsequent fibrosis. Biomarkers of function include serum creatinine levels for kidney transplants, pulmonary function tests for lung transplants, and bile or specific enzyme levels for liver transplants.
- HLA restriction
-
An analytical condition in which association tests are confined to subgroups that share common HLA alleles.
- Lead SNP
-
The single-nucleotide polymorphism (SNP) within any given locus in a genome-wide association study that has the strongest statistical significance.
- Interstitial fibrosis and tubular atrophy
-
(IFTA). Historically called chronic allograft nephropathy, it is the most common cause of long-term renal graft failure and is characterizedby the gradual deterioration of graft function.
- Pharmacokinetics
-
The study of how an organism affects a pharmaceutical agent, one aspect of which is the metabolism of the drug. The levels of immunosuppressive drugs are highly affected by individual variability in specific metabolizing enzymes.
- Acute pyelonephritis
-
A bacterial infection of the kidney. Individuals who are taking immunosuppressive medication are at an increased risk of developing this condition.
- Xenografts
-
Grafts from one species to a different species, such as in the case of heart valve replacement, which often involves the transplantation of heart valves from pigs into humans.
- Sirtuins
-
A class of proteins implicated in processes such as ageing, apoptosis and inflammation.
Rights and permissions
About this article
Cite this article
Yang, J., Sarwal, M. Transplant genetics and genomics. Nat Rev Genet 18, 309–326 (2017). https://doi.org/10.1038/nrg.2017.12
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2017.12
This article is cited by
-
A practical approach to the genomics of kidney disorders
Pediatric Nephrology (2022)
-
Liquid biopsies: donor-derived cell-free DNA for the detection of kidney allograft injury
Nature Reviews Nephrology (2021)
-
An accessible insight into genetic findings for transplantation recipients with suspected genetic kidney disease
npj Genomic Medicine (2021)
-
HLA and kidney disease: from associations to mechanisms
Nature Reviews Nephrology (2018)