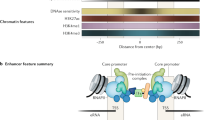
Key Points
-
Enhancers are distal regulatory genomic elements that determine spatiotemporal and quantitative gene transcription programmes in response to developmental or environmental cues. Global transcriptomic and (epi)genomic advances have found that enhancers are pervasively transcribed into non-coding RNAs (ncRNAs), which are named enhancer RNAs (eRNAs).
-
The expression levels of eRNAs are highly correlated with the activity of the functional enhancers, both in developmental and rapid signal-regulated transcriptional programmes. eRNAs bear several common and unique features compared to other long ncRNAs or coding mRNAs.
-
Heterogeneity of eRNAs exists in terms of both transcriptional processes and RNA properties, which might be linked to their functional diversity. There is evidence supporting a functional role of both the eRNA transcripts per se, and the act of transcription, in the activation of target coding genes. However, for the large majority of eRNAs in the mammalian genome, evidence of their functional roles is still limited.
-
Mechanisms underlying the roles of specific eRNAs include modulating the chromatin accessibility of cognate promoters, stabilizing enhancer–promoter interactions, trapping transcription factors on genomic sites, and regulating the RNA polymerase II pause release at cognate promoters. The act of enhancer transcription may facilitate histone modifications, remodel large areas of chromatin and induce transcriptional interference.
-
Enhancer transcription may take part in generating several important biological phenomena, including transcription-dependent R-loops, genomic mutations and instability at enhancers, and potentially facilitate new gene birth during evolution.
-
Further evidence is needed to fully understand the functions of different categories of enhancers as transcription units and the roles of eRNAs per se, in gene regulation, development and disease.
Abstract
Networks of regulatory enhancers dictate distinct cell identities and cellular responses to diverse signals by instructing precise spatiotemporal patterns of gene expression. However, 35 years after their discovery, enhancer functions and mechanisms remain incompletely understood. Intriguingly, recent evidence suggests that many, if not all, functional enhancers are themselves transcription units, generating non-coding enhancer RNAs. This observation provides a fundamental insight into the inter-regulation between enhancers and promoters, which can both act as transcription units; it also raises crucial questions regarding the potential biological roles of the enhancer transcription process and non-coding enhancer RNAs. Here, we review research progress in this field and discuss several important, unresolved questions regarding the roles and mechanisms of enhancers in gene regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). References 2 and 3 report the most comprehensive transcriptomic profiling to date of eRNAs in humans.
Bulger, M. & Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011).
Levine, M. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763 (2010).
Plank, J. L. & Dean, A. Enhancer function: mechanistic and genome-wide insights come together. Mol. Cell 55, 5–14 (2014).
Cech, T. R. & Steitz, J. A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94 (2014).
Quinn, J. J. & Chang, H. Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62 (2015).
Goff, L. A. & Rinn, J. L. Linking RNA biology to lncRNAs. Genome Res. 25, 1456–1465 (2015).
Calo, E. & Wysocka, J. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49, 825–837 (2013).
Shlyueva, D., Stampfel, G. & Stark, A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272–286 (2014).
Rivera, C. M. & Ren, B. Mapping human epigenomes. Cell 155, 39–55 (2013).
Blackwood, E. M. & Kadonaga, J. T. Going the distance: a current view of enhancer action. Science 281, 60–63 (1998).
Banerji, J., Rusconi, S. & Schaffner, W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308 (1981).
Moreau, P. et al. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 9, 6047–6068 (1981).
Banerji, J., Olson, L. & Schaffner, W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33, 729–740 (1983).
Gillies, S. D., Morrison, S. L., Oi, V. T. & Tonegawa, S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33, 717–728 (1983).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Heintzman, N. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Spitz, F. & Furlong, E. E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012).
Visel, A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009). References 19 and 21 set the foundation for the large-scale identification of potentially active enhancers using histone H3K4 methylation marks together with p300.
Bonn, S. et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44, 148–156 (2012).
Pekowska, A. et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 30, 4198–4210 (2011).
Ashe, H. L., Monks, J., Wijgerde, M., Fraser, P. & Proudfoot, N. J. Intergenic transcription and transinduction of the human β-globin locus. Genes Dev. 11, 2494–2509 (1997).
Tuan, D., Kong, S. & Hu, K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc. Natl Acad. Sci. USA 89, 11219–11223 (1992). This study first identified the presence of enhancer transcription in mammalian cells.
Masternak, K., Peyraud, N., Krawczyk, M., Barras, E. & Reith, W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 4, 132–137 (2003).
Rogan, D. F. et al. Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc. Natl Acad. Sci. USA 101, 2446–2451 (2004).
Lipshitz, H. D., Peattie, D. A. & Hogness, D. S. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1, 307–322 (1987). This study identified the first intergenic non-coding transcripts in Drosophila melanogaster.
Feng, J. et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484 (2006).
Cheng, J. et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154 (2005).
Carroll, J. S. et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38, 1289–1297 (2006).
De Santa, F. et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8, e1000384 (2010).
Kim, T.-K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010). References 32 and 33 report the first two genome-wide efforts to identify pervasive RNA production from putative enhancer regions.
Hah, N. et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634 (2011).
Wang, D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011).
Allen, M. A. et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife 3, e02200 (2013).
Alvarez-Dominguez, J. R. et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood 123, 570–581 (2014).
Arner, E. et al. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347, 1010–1014 (2015). This study provides evidence that a subset of enhancers is temporally the 'leading' transcription units, representing the earliest activation events in response to stimulus.
Fang, B. et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152 (2014).
Hsieh, C.-L. et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl Acad. Sci. USA 111, 7319–7324 (2014).
Iiott, N. E. et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 5, 3979 (2013).
Lai, F., Gardini, A., Zhang, A. & Shiekhattar, R. Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403 (2015).
Lam, M. et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498, 511–515 (2013).
Li, W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013).
Melo, C. et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 49, 524–535 (2013).
Meng, F. L. et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell 159, 1538–1548 (2014).
Mousavi, K. et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 51, 606–617 (2013).
Ounzain, S. et al. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell Cardiol. 76, 55–70 (2014).
Pefanis, E. et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789 (2015). This study identifies a role of RNA exosome subunits in modulating eRNA transcription and stability and the related R-loop formation on some specific enhancers.
Pnueli, L., Rudnizky, S., Yosefzon, Y. & Melamed, P. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin α-subunit gene. Proc. Natl Acad. Sci. USA 112, 4369–4374 (2015).
Sigova, A. et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl Acad. Sci. USA 110, 2876–2881 (2013).
Skowronska-Krawczyk, D. et al. Required enhancer-matrin-3 network interactions for a homeodomain transcription program. Nature 514, 257–261 (2014).
Wu, H. et al. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS Genet. 10, e1004610 (2014). The first in vivo study to use transgenic mice embryos to confirm the correlation between eRNA transcription and the likelihood of that enhancer being functional.
Blum, R., Vethantham, V., Bowman, C., Rudnicki, M. & Dynlacht, B. D. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 26, 2763–2779 (2012).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Parker, S. C. et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl Acad. Sci. USA 110, 17921–17926 (2013). References 55 and 56 first coined the concept of super and stretch enhancers, respectively, to denote the widespread phenomenon of clustered enhancers in mammalian cells.
Barolo, S. Shadow enhancers: frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 34, 135–141 (2012).
Montavon, T. et al. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Gerstein, M. B. et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787 (2010).
Junion, G. et al. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148, 473–486 (2012).
Liu, Z. et al. Enhancer activation requires trans-recruitment of a mega transcription factor complex. Cell 159, 358–373 (2014).
Pott, S. & Lieb, J. D. What are super-enhancers? Nat. Genet. 47, 8–12 (2015).
Hah, N. et al. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc. Natl Acad. Sci. USA 112, E297–E302 (2015).
Hah, N., Murakami, S., Nagari, A., Danko, C. & Kraus, W. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 23, 1210–1223 (2013).
Melgar, M. F., Collins, F. S. & Sethupathy, P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 12, R113 (2010). References 32–35 and 67 are the first of several reports correlating eRNA transcription with enhancer activation on a genome-wide scale.
Zhu, Y. et al. Predicting enhancer transcription and activity from chromatin modifications. Nucleic Acids Res. 41, 10032–10043 (2013).
Pulakanti, K. et al. Enhancer transcribed RNAs arise from hypomethylated, Tet-occupied genomic regions. Epigenetics 8, 1303–1320 (2013).
Schlesinger, F., Smith, A. D., Gingeras, T. R., Hannon, G. J. & Hodges, E. De novo DNA demethylation and noncoding transcription define active intergenic regulatory elements. Genome Res. 23, 1601–1614 (2013).
Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012).
Andersson, R. et al. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat. Commun. 5, 5336 (2014).
Core, L. J. et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320 (2014). References 72 and 73 thoroughly analyse multiple features of transcriptional events from enhancers and promoters.
Stasevich, T. J. et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516, 272–275 (2014).
Kundu, T. K. et al. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6, 551–561 (2000).
Lenhard, B., Sandelin, A. & Carninci, P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 13, 233–245 (2012).
Preker, P. et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 39, 7179–7193 (2011).
Scruggs, B. S. et al. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol. Cell 58, 1101–1112 (2015). This paper found that the initiation sites for some promoter upstream transcripts carry similar epigenomic features to enhancers.
Lubas, M. et al. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 10, 178–192 (2015). This study reported that eRNAs are broadly bound by RNA-binding protein 7 (RBM7), which is responsible for loading the RNA exosome complex onto the nascent transcripts.
Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008).
Carroll, J. S. et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005).
Kaikkonen, M. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325 (2013).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Bieda, M., Xu, X., Singer, M. A., Green, R. & Farnham, P. J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16, 595–605 (2006).
Boyle, A. P. et al. Comparative analysis of regulatory information and circuits across distant species. Nature 512, 453–456 (2014).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Sigova, A. A. et al. Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981 (2015). This paper supports a hypothesis that a general role of ncRNAs, including eRNAs, is to assist the binding of transcription factors, such as YY1, to the regulatory elements.
Hatzis, P. & Talianidis, I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10, 1467–1477 (2002).
Koch, F. et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 18, 956–963 (2011).
Muller-McNicoll, M. & Neugebauer, K. M. Good cap/bad cap: how the cap-binding complex determines RNA fate. Nat. Struct. Mol. Biol. 21, 9–12 (2014).
Bentley, D. L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 (2014).
Almada, A. E., Wu, X., Kriz, A. J., Burge, C. B. & Sharp, P. A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013).
Ntini, E. et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nature Struct. Mol. Biol. 20, 923–928 (2013).
Kowalczyk, M. S. et al. Intragenic enhancers act as alternative promoters. Mol. Cell 45, 447–458 (2012).
Kanno, T. K. et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol. 21, 1047–1057 (2014).
Keogh, M. C. et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123, 593–605 (2005).
de Almeida, S. F. et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 18, 977–983 (2011).
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Bieberstein, N. I., Carrillo Oesterreich, F., Straube, K. & Neugebauer, K. M. First exon length controls active chromatin signatures and transcription. Cell Rep. 2, 62–68 (2012).
Sims, R. J. et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28, 665–676 (2007).
Clouaire, T. et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 26, 1714–1728 (2012).
Baillat, D. et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276 (2005).
Austenaa, L. M. et al. Transcription of mammalian cis-regulatory elements is restrained by actively enforced early termination. Mol. Cell 60, 460–474 (2015). References 42 and 103 unravel the regulatory roles of the Integrator complex and WDR82 in eRNA transcriptional termination.
Descostes, N. et al. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. eLife 3, e02105 (2013). This study identifies the enrichment of Tyr1p at active enhancers.
Hsin, J. P., Li, W., Hoque, M., Tian, B. & Manley, J. L. RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. eLife 3, e02112 (2014).
Mayer, A. et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336, 1723–1725 (2012).
Aguilo, F. et al. Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1a. Cell Rep. 14, 1–14 (2016). This paper reports the deposition of the 5mC mark on some eRNAs.
Ling, J., Baibakov, B., Pi, W., Emerson, B. M. & Tuan, D. The HS2 enhancer of the β-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J. Mol. Biol. 350, 883–896 (2005).
Kim, Y. W., Lee, S., Yun, J. & Kim, A. Chromatin looping and eRNA transcription precede the transcriptional activation of gene in the β-globin locus. Biosci. Rep. 35, e00179 (2015).
Schaukowitch, K. et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56, 29–42 (2014). The first report to reveal that some eRNAs act as decoys to sequester some transcriptionally repressive complexes from target gene promoters.
Ragoczy, T., Bender, M. A., Telling, A., Byron, R. & Groudine, M. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 20, 1447–1457 (2006).
Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012).
Struhl, K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14, 103–105 (2007).
Banerjee, A. R., Kim, Y. J. & Kim, T. H. A novel virus-inducible enhancer of the interferon-β gene with tightly linked promoter and enhancer activities. Nucleic Acids Res. 42, 12537–12554 (2014).
Lai, F. et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013). References 40, 44 and 115 are the first papers to show a role of interrogated eRNAs in facilitating the formation of enhancer–promoter looping.
Maruyama, A., Mimura, J. & Itoh, K. Non-coding RNA derived from the region adjacent to the human HO-1 E2 enhancer selectively regulates HO-1 gene induction by modulating Pol II binding. Nucleic Acids Res. 42, 13599–13614 (2014).
Orom, U. A. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58 (2010). References 40, 47 and 117 suggest the potential trans roles of some eRNAs.References 41, 43–45, 47, 115 and 117 first reveal the effects of eRNA knockdown on the expression of cognate coding genes.
Villar, D. et al. Enhancer evolution across 20 mammalian species. Cell 160, 554–566 (2015).
Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012). This paper reports that some promoters act like enhancers in reporter assays.
Herbert, K. M., Greenleaf, W. J. & Block, S. M. Single-molecule studies of RNA polymerase: motoring along. Annu. Rev. Biochem. 77, 149–176 (2008).
Gribnau, J., Diderich, K., Pruzina, S., Calzolari, R. & Fraser, P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5, 377–386 (2000).
Ho, Y., Elefant, F., Liebhaber, S. A. & Cooke, N. E. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 23, 365–375 (2006).
Ling, J. et al. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J. Biol. Chem. 279, 51704–51713 (2004). References 82, 122 and 123 are three important reports that suggest roles for the enhancer transcriptional process.
Onodera, C. S. et al. Gene isoform specificity through enhancer-associated antisense transcription. PLoS ONE 7, e43511 (2012).
Shechner, D. M., Hacisuleyman, E., Younger, S. T. & Rinn, J. L. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods 12, 664–670 (2015).
Trimarchi, T. et al. Genome-wide mapping and characterization of notch-regulated long noncoding RNAs in acute leukemia. Cell 158, 593–606 (2014).
Wang, K. C. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011). References 29 and 127 are the first two reports on the activating roles of lncRNAs.
Xiang, J. F. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 24, 513–531 (2014).
Cifuentes-Rojas, C., Hernandez, A. J., Sarma, K. & Lee, J. T. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell 55, 171–185 (2014).
Davidovich, C., Zheng, L., Goodrich, K. J. & Cech, T. R. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 20, 1250–1257 (2013).
Kaneko, S., Son, J., Shen, S. S., Reinberg, D. & Bonasio, R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1258–1264 (2013).
Kung, J. T. et al. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol. Cell 57, 361–375 (2015).
Chu, C. et al. Systematic discovery of xist RNA binding proteins. Cell 161, 404–416 (2015).
McHugh, C. A. et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015).
Minajigi, A. et al. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, aab2276 (2015).
Zhang, Y. et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504, 306–310 (2013).
Liu, Z. et al. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife 3, e04236 (2014).
Caudron-Herger, M., Cook, P. R., Rippe, K. & Papantonis, A. Dissecting the nascent human transcriptome by analysing the RNA content of transcription factories. Nucleic Acids Res. 43, e95 (2015).
Mao, Y. S., Sunwoo, H., Zhang, B. & Spector, D. L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13, 95–101 (2011).
Santos-Pereira, J. M. & Aguilera, A. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 16, 583–597 (2015).
Puc, J. et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell 160, 367–380 (2015).
Qian, J. et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell 159, 1524–1537 (2014).
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I. & Chédin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015).
Tautz, D. & Domazet-Loso, T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 12, 692–702 (2011).
Young, R. S. et al. The frequent evolutionary birth and death of functional promoters in mouse and human. Genome Res. 25, 1546–1557 (2015).
Wu, X. & Sharp, P. A. Divergent transcription: a driving force for new gene origination? Cell 155, 990–996 (2013).
Kim, T. K. & Shiekhattar, R. Architectural and functional commonalities between enhancers and promoters. Cell 162, 948–959 (2015).
Hnisz, D. et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell 58, 362–370 (2015).
Yao, P. et al. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat. Neurosci. 18, 1168–1174 (2015).
Farh, K. K. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2014). References 3, 145 and 152 show that the enhancers enriched in disease-associated genetic variations display transcription of eRNAs.
Engreitz, J. M. et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 159, 188–199 (2014).
Wang, Q., Carroll, J. S. & Brown, M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19, 631–642 (2005).
de Wit, E. & de Laat, W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 (2012).
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Hughes, J. R. et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212 (2014).
Mifsud, B. et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 47, 598–606 (2015).
Pombo, A. & Dillon, N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 16, 245–257 (2015).
Ghavi-Helm, Y. et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96–100 (2014).
Fullwood, M. J. et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 (2009).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Li, L. et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol. Cell 58, 216–231 (2015).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Williamson, I. et al. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 28, 2778–2791 (2014).
Vucicevic, D., Corradin, O., Ntini, E., Scacheri, P. C. & Orom, U. A. Long ncRNA expression associates with tissue-specific enhancers. Cell Cycle 14, 253–260 (2015).
Cheng, J. H., Pan, D. Z., Tsai, Z. T. & Tsai, H. K. Genome-wide analysis of enhancer RNA in gene regulation across 12 mouse tissues. Sci. Rep. 5, 12648 (2015).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Li, W. et al. Condensin I and II complexes license full estrogen receptor α-dependent enhancer activation. Mol. Cell 59, 188–202 (2015).
Shen, L. et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706 (2013).
Song, C. X. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 (2013).
Hon, G. C. et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol. Cell 56, 286–297 (2014).
Ceballos-Chavez, M. et al. The chromatin Remodeler CHD8 is required for activation of progesterone receptor-dependent enhancers. PLoS Genet. 11, e1005174 (2015).
Wang, X. et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130 (2008).
Allo, M. et al. Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc. Natl Acad. Sci. USA 111, 15622–15629 (2014).
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008).
Kwak, H., Fuda, N. J., Core, L. J. & Lis, J. T. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339, 950–953 (2013).
Kruesi, W. S., Core, L. J., Waters, C. T., Lis, J. T. & Meyer, B. J. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2, e00808 (2013).
Magnuson, B. et al. Identifying transcription start sites and active enhancer elements using BruUV-seq. Sci. Rep. 5, 17978 (2015).
Mayer, A. et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 (2015).
Nojima, T. et al. Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161, 526–540 (2015).
Mercer, T. R. et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 30, 99–104 (2012).
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E. & Chang, H. Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678 (2011).
Acknowledgements
The authors thank M. Chuang for her helpful comments on the manuscript. This work was supported by the US Department of Defense breast cancer research programme postdoctoral fellowships (BC110381 to W.L. and BC103858 to D.N.) and grants from the US National Institutes of Health to M.G.R. M.G.R. is an investigator with the Howard Hughes Medical Institute. The authors apologize for not being able to cite many important other publications owing to the imposed limit.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Non-coding
-
DNA regions or RNA transcripts that do not code for proteins.
- Long non-coding RNAs
-
(lncRNAs). Non-coding RNAs that are longer than 200 nucleotides.
- Hypersensitivity
-
Chromatin regions such as enhancers and other regulatory elements often display extra sensitivity to DNase treatment, reflecting the openness of the regions.
- Locus control regions
-
(LCRs). Genomic elements that elevate expression of linked genes through long-range regulation, in a copy number-dependent and tissue-specific manner. A well-studied example is the LCR upstream of the β-globin gene in erythroid cells.
- Super enhancers
-
A group of active enhancers densely clustered in a ~10–30 kb region, highly associated with cell identity genes and disease-associated genomic variations. Also known as stretch enhancers.
- Shadow enhancers
-
A term coined by Michael Levine and colleagues to describe the phenomenon of having another enhancer in the vicinity (or sometimes scattered across larger chromosomal domains) in addition to a primary enhancer to control the expression pattern of an important developmental gene; their biological roles are still under investigation, but, at least in some cases, shadow enhancers act to confer phenotypic robustness under environmental and genetic variability.
- Regulatory archipelagos
-
A term denoting the presence of multiple enhancers in the Hox gene loci during limb development, with each of them playing quantitative or qualitative roles for Hox gene transcription.
- Highly occupied target regions
-
(HOT regions; also known as hotspots). Genomic regions that associate with multiple transcription factors, either simultaneously or sequentially, and that are usually uncovered by chromatin immunoprecipitation followed by sequencing. They are identified in multiple organisms and cell types.
- TF collective or MegaTrans enhancer
-
Highly active enhancers that are bound by multiple transcription factors simultaneously (similar to the definition of HOT regions and super enhancers). However, these two terms have been used to describe a situation in which one (MegaTrans) or several (TF collective) major transcription factors bind target enhancers in cis (that is, direct association through a specific DNA motif), which then act to tether other transcription factors in trans (that is, bind the major transcription factor through protein–protein interaction).
- Promoter upstream transcripts
-
(PROMPTs). Primary transcripts that are generated pervasively from gene promoters but are transcribed in the opposite direction from the sense strand (that is, mRNAs). PROMPTs generally display low stability, lack of splicing and polyadenylation; very similar to enhancer RNAs in many aspects. Also known as upstream antisense RNAs.
- General transcription factors
-
Transcription factors that work together with RNA polymerase II to form the pre-initiation complex at transcription start sites to initiate transcription. They consist of TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH.
- Bidirectional transcripts
-
The two transcripts initially observed to be generated by some coding gene promoters that go to either a sense or an antisense direction, which produces the mRNAs and the promoter upstream transcripts, respectively. A similar phenomenon is now observed to exist for some enhancers.
- DRB
-
(5,6-dichloro-1-β-D-ribofuranosylbenzimidazole). An adenosine analogue that acts as an inhibitor of cyclin-dependent kinases needed for efficient RNA polymerase II elongation.
- RNA exosome
-
A multi-protein complex capable of degrading various types of RNA molecules in the cytoplasm, nucleus and the nucleolus; it bears both exo- and endo-ribonucleolytic functions in eukaryotes.
- RNAPII carboxy-terminal domain
-
(RNAPII CTD). An evolutionary conserved tandem repeat of heptapeptides Y1S2P3T4S5P6S7 that is present in the C terminus of RPB1, the largest subunit of RNA polymerase II (RNAPII).
- Small nuclear RNAs
-
A class of small RNAs in the nucleus of eukaryotic cells that have been found to largely take part in regulating splicing (for example, U1 and U2 RNAs) and occasionally in transcriptional control of RNA polymerase II (for example, 7SK RNA).
- Enhancer–promoter inter-regulation
-
A hypothesis in which active enhancers affect promoter expression, and some promoters also control enhancer transcription.
- Transcriptional noise
-
A term used to denote non-productive and perhaps random transcriptional activity of an RNA polymerase on a DNA template, which is proposed to take place due to open chromatin regions.
- De novo enhancers
-
A group of enhancers that were not marked by any epigenomic marks during cell lineage determination, but rather are generated acutely in a mature cell type after treatment with acute stimulation; they are also known as latent enhancers.
- NELF complex
-
(Negative elongation factor complex). A four-subunit complex consisting of NELF-A, NELF-B, NELF-E and either NELF-C or NELF-D. As denoted by the name, it negatively affects transcription by RNA polymerase II.
- Myogenic differentiation 1
-
(MyoD1). A gene encoding a key transcription factor that promotes muscle-specific gene transcription programmes that are required for myogenic determination.
- Genomic variations
-
The varied DNA sequences in alleles of certain genes carried by individuals within and among populations, which may or may not result in phenotypic variations.
- R-loops
-
RNA/DNA hybrid structures in the genome, in which nascent RNA binds the transcribing DNA strand through sequence complementarity, leaving the non-bound single-strand DNA displaced and prone to damage. Once thought to be transcriptional by-products, R-loops have now been found to be involved in the regulation of gene expression, epigenetic modifications, DNA replication and genome stability.
- Somatic hypermutation
-
A biological process mainly conducted by activation-induced cytidine deaminase in activated B cells, in which the immunoglobulin genes are highly mutated to generate a library of diversified antibodies.
- Class switch recombination
-
A biological mechanism enabling B cells to switch their production of immunoglobulin from one type to another (for example, IgM to IgG), which involves an activation-induced cytidine deaminase-mediated specific DNA double-strand break and recombination.
- Single nucleotide polymorphisms
-
(SNPs). Genomic variations that involve a single nucleotide.
Rights and permissions
About this article
Cite this article
Li, W., Notani, D. & Rosenfeld, M. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17, 207–223 (2016). https://doi.org/10.1038/nrg.2016.4
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2016.4
This article is cited by
-
Bivalent activity of super-enhancer RNA LINC02454 controls 3D chromatin structure and regulates glioma sensitivity to temozolomide
Cell Death & Disease (2024)
-
Mapping the epigenomic landscape of human monocytes following innate immune activation reveals context-specific mechanisms driving endotoxin tolerance
BMC Genomics (2023)
-
Decoding enhancer complexity with machine learning and high-throughput discovery
Genome Biology (2023)
-
m6A methylation of super-enhancer RNAs facilitates chromatin accessibility
Nature Genetics (2023)
-
JUN-induced super-enhancer RNA forms R-loop to promote nasopharyngeal carcinoma metastasis
Cell Death & Disease (2023)