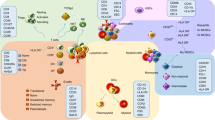
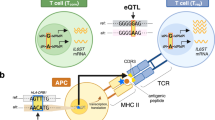
Key Points
-
Autoimmune diseases affect approximately 8–9% of the world population. There are no cures and only limited therapies exist to treat symptoms.
-
Hundreds of genomic susceptibility loci have been discovered but, despite progress in fine-mapping causal genes and variant identification, the genetic mechanisms that trigger autoimmunity are largely unknown.
-
Most risk variants that are likely to be causal are in non-coding sequences, particularly in regulatory regions known to be active in immune cell types; it is therefore crucial to understand how enhancers and their targets affect gene regulation and immune function.
-
As a wider compendium of transcriptomic and epigenomic profiles for human immune cell types and cell states is built, we will be able to further pinpoint the conditions under which most risk variants may be exerting their effect.
-
Profiling of immunophenotypes, such as signalling response, immune cell abundances and serum cytokine levels (among others), in thousands of individuals may help identify the key immune functions and interactions involved in autoimmunity risk.
-
When the key immunophenotypes have been identified, longitudinal studies will be essential for revealing the causal relationships among intermediate phenotypes and elucidating the sequence of events that lead to autoimmunity.
Abstract
Genome-wide strategies have driven the discovery of more than 300 susceptibility loci for autoimmune diseases. However, for almost all loci, understanding of the mechanisms leading to autoimmunity remains limited, and most variants that are likely to be causal are in non-coding regions of the genome. A critical next step will be to identify the in vivo and ex vivo immunophenotypes that are affected by risk variants. To do this, key cell types and cell states that are implicated in autoimmune diseases will need to be defined. Functional genomic annotations from these cell types and states can then be used to resolve candidate genes and causal variants. Together with longitudinal studies, this approach may yield pivotal insights into how autoimmunity is triggered.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mackay, I. R. Travels and travails of autoimmunity: a historical journey from discovery to rediscovery. Autoimmun. Rev. 9, A251–A258 (2010).
Hayter, S. M. & Cook, M. C. Updated assessment of the prevalence, spectrum and case definition of autoimmune disease. Autoimmun. Rev. 11, 754–765 (2012).
Cooper, G. S., Bynum, M. L. K. & Somers, E. C. Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun. 33, 197–207 (2009). This is a good epidemiology review with prevalence estimates for autoimmune diseases.
Walsh, S. J. & Rau, L. M. Autoimmune diseases: a leading cause of death among young and middle-aged women in the United States. Am. J. Public Health 90, 1463–1466 (2000).
Thomas, S. L., Griffiths, C., Smeeth, L., Rooney, C. & Hall, A. J. Burden of mortality associated with autoimmune diseases among females in the United Kingdom. Am. J. Public Health 100, 2279–2287 (2010).
Gilkeson, G. et al. The United States to Africa lupus prevalence gradient revisited. Lupus 20, 1095–1103 (2011).
Davis, M. M. A prescription for human immunology. Immunity 29, 835–838 (2008).
Cotsapas, C. et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 7, e1002254 (2011). This study uses GWAS data to show extensive sharing of genetic risk for seven autoimmune diseases beyond the MHC and PTPN22 loci.
Bogdanos, D. P. et al. Twin studies in autoimmune disease: genetics, gender and environment. J. Autoimmun. 38, J156–J169 (2012).
Willer, C. J. et al. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc. Natl Acad. Sci. USA 100, 12877–12882 (2003).
Hawkes, C. H. & Macgregor, A. J. Twin studies and the heritability of MS: a conclusion. Mult. Scler. 15, 661–667 (2009).
Elder, J. T. et al. The genetics of psoriasis. Arch. Dermatol. 130, 216–224 (1994).
Satsangi, J. et al. Contribution of genes of the major histocompatibility complex to susceptibility and disease phenotype in inflammatory bowel disease. Lancet 347, 1212–1217 (1996).
Vyse, T. J. & Todd, J. A. Genetic analysis of autoimmune disease. Cell 85, 311–318 (1996).
Raychaudhuri, S. Recent advances in the genetics of rheumatoid arthritis. Curr. Opin. Rheumatol. 22, 109–118 (2010).
Criswell, L. A. et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am. J. Hum. Genet. 76, 561–571 (2005).
Eaton, W. W., Rose, N. R., Kalaydjian, A., Pedersen, M. G. & Mortensen, P. B. Epidemiology of autoimmune diseases in Denmark. J. Autoimmun. 29, 1–9 (2007).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Rich, S. S., Weitkamp, L. R. & Barbosa, J. Genetic heterogeneity of insulin-dependent (type I) diabetes mellitus: evidence from a study of extended haplotypes. Am. J. Hum. Genet. 36, 1015–1023 (1984).
Gaffney, P. M. et al. A genome-wide search for susceptibility genes in human systemic lupus erythematosus sib-pair families. Proc. Natl Acad. Sci. USA 95, 14875–14879 (1998).
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's's disease. Nature 411, 599–603 (2001).
Rioux, J. D. et al. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am. J. Hum. Genet. 66, 1863–1870 (2000).
Hu, X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47, 898–905 (2015).
The MHC Sequencing Consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature 401, 921–923 (1999).
Tomfohrde, J. et al. Gene for familial psoriasis susceptibility mapped to the distal end of human chromosome 17q. Science 264, 1141–1145 (1994).
Jordan, C. T. et al. PSORS2 is due to mutations in CARD14. Am. J. Hum. Genet. 90, 784–795 (2012).
Plenge, R. M. et al. Replication of putative candidate-gene associations with rheumatoid arthritis in>4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am. J. Hum. Genet. 77, 1044–1060 (2005).
Begovich, A. B. et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 75, 330–337 (2004).
Velaga, M. R. et al. The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves' disease. J. Clin. Endocrinol. Metab. 89, 5862–5865 (2004).
Kyogoku, C. et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am. J. Hum. Genet. 75, 504–507 (2004).
Bottini, N. et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 36, 337–338 (2004).
Rawlings, D. J., Dai, X. & Buckner, J. H. The role of PTPN22 risk variant in the development of autoimmunity: finding common ground between mouse and human. J. Immunol. 194, 2977–2984 (2015).
Nisticò, L., Buzzetti, R., Pritchard, L. A. et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with type 1 diabetes. Hum. Molec. Genet. 5, 1075–1080 (1996)
Petukhova, L. et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature 466, 113–117 (2010).
Fortune, M. D. et al. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat. Genet. 47, 839–846 (2015).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Jostins, L. et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). This large meta-analysis of GWAS reveals the greatest number of susceptibility loci for IBD.
Farh, K. K.-H. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015). This study fine-maps autoimmune disease variants and integrates them with immune enhancer and promoter marks, as well as with blood eQTLs.
Li, Y. R. et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat. Med. 21, 1018–1027 (2015).
Barratt, B. J. et al. Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes 53, 1884–1889 (2004).
Onengut-Gumuscu, S. et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 47, 381–386 (2015). This paper increases the resolution of the SNPs most associated with T1DM, highlights the genetic relationships between T1DM and other autoimmune diseases, and uses functional annotation to implicate specific cell types and regulatory DNA (enhancers).
Parkes, M., Cortes, A., van Heel, D. A. & Brown, M. A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 14, 661–673 (2013).
Ramos, P. S., Shedlock, A. M. & Langefeld, C. D. Genetics of autoimmune diseases: insights from population genetics. J. Hum. Genet. 60, 657–664 (2015).
Raj, T. et al. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am. J. Hum. Genet. 92, 517–529 (2013).
Cagliani, R. et al. Crohn's's disease loci are common targets of protozoa-driven selection. Mol. Biol. Evol. 30, 1077–1087 (2013).
Stastny, P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N. Engl. J. Med. 298, 869–871 (1978).
Gregersen, P. K., Silver, J. & Winchester, R. J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 30, 1205–1213 (1987). This study describes the shared epitope hypothesis as a common thread for MHC effects.
Raychaudhuri, S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 (2012). This study shows that the MHC effects for RA resolve into specific amino acids in the peptide-binding grooves of MHC molecules.
Okada, Y. et al. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum. Mol. Genet. 23, 6916–6926 (2014).
Reynolds, R. J. et al. HLA-DRB1-associated rheumatoid arthritis risk at multiple levels in African Americans: hierarchical classification systems, amino acid positions, and residues. Arthritis Rheumatol. 66, 3274–3282 (2014).
Liu, J. Z. et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986 (2015).
Han, B. et al. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am. J. Hum. Genet. 94, 522–532 (2014).
Foo, J. N. et al. Coding variants at hexa-allelic amino acid 13 of HLA-DRB1 explain independent SNP associations with follicular lymphoma risk. Am. J. Hum. Genet. 93, 167–172 (2013).
Zheng, X. et al. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 14, 192–200 (2014).
Leslie, S., Donnelly, P. & McVean, G. A statistical method for predicting classical HLA alleles from SNP data. Am. J. Hum. Genet. 82, 48–56 (2008).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE 8, e64683 (2013).
International HIV Controllers Study. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330, 1551–1557 (2010).
Okada, Y. et al. Construction of a population-specific HLA imputation reference panel and its application to Graves' disease risk in Japanese. Nat. Genet. 47, 798–802 (2015).
Warren, R. L. et al. Derivation of HLA types from shotgun sequence datasets. Genome Med. 4, 95 (2012).
Liu, C. et al. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 41, e142 (2013).
Bai, Y., Ni, M., Cooper, B., Wei, Y. & Fury, W. Inference of high resolution HLA types using genome-wide RNA or DNA sequencing reads. BMC Genomics 15, 325 (2014).
Huang, Y. et al. HLAreporter: a tool for HLA typing from next generation sequencing data. Genome Med. 7, 25 (2015).
Dilthey, A., Cox, C., Iqbal, Z., Nelson, M. R. & McVean, G. Improved genome inference in the MHC using a population reference graph. Nat. Genet. 47, 682–688 (2015).
Horton, R. et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics 60, 1–18 (2008).
Holdsworth, R. et al. The HLA dictionary 2008: a summary of HLA-A, -B, -C, -DRB1/3/4/5, and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Tissue Antigens 73, 95–170 (2009).
Cereb, N., Kim, H. R., Ryu, J. & Yang, S. Y. Advances in DNA sequencing technologies for high resolution HLA typing. Hum. Immunol. 76, 923–927 (2015).
Cortes, A. & Brown, M. A. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 13, 101 (2011).
Trynka, G. et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 43, 1193–1201 (2011).
Huang, H. et al. Association mapping of inflammatory bowel disease loci to single variant resolution. bioRxiv http://dx.doi.org/10.1101/028688 (2015).
Cooper, J. D. et al. Seven newly identified loci for autoimmune thyroid disease. Hum. Mol. Genet. 21, 5202–5208 (2012).
International Genetics of Ankylosing Spondylitis Consortium (IGAS). Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45, 730–738 (2013).
Hinks, A. et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat. Genet. 45, 664–669 (2013).
Tsoi, L. C. et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 44, 1341–1348 (2012).
Eyre, S. et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet. 44, 1336–1340 (2012).
Wei, Z. et al. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am. J. Hum. Genet. 92, 1008–1012 (2013).
Vilhjálmsson, B. J. et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 97, 576–592 (2015).
Peloso, G. M. et al. Phenotypic extremes in rare variant study designs. Eur. J. Hum. Genet. http://dx.doi.org/10.1038/ejhg.2015.197 (2015).
MacArthur, D. G. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335, 823–828 (2012).
Hunt, K. A. et al. Rare and functional SIAE variants are not associated with autoimmune disease risk in up to 66,924 individuals of European ancestry. Nat. Genet. 44, 3–5 (2012).
Dickson, S. P., Wang, K., Krantz, I., Hakonarson, H. & Goldstein, D. B. Rare variants create synthetic genome-wide associations. PLoS Biol. 8, e1000294 (2010).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
Rivas, M. A. et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 43, 1066–1073 (2011). One of the first deep re-sequencing studies to find rare variants involved in IBD, which also assesses their functional consequences.
Diogo, D. et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS ONE 10, e0122271 (2015).
Nejentsev, S., Walker, N., Riches, D., Egholm, M. & Todd, J. A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324, 387–389 (2009).
Meylan, E., Tschopp, J. & Karin, M. Intracellular pattern recognition receptors in the host response. Nature 442, 39–44 (2006).
Rice, G. I. et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 46, 503–509 (2014).
Beaudoin, M. et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 9, e1003723 (2013).
Bang, S.-Y. et al. Targeted exon sequencing fails to identify rare coding variants with large effect in rheumatoid arthritis. Arthritis Res. Ther. 16, 447 (2014).
Hunt, K. A. et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature 498, 232–235 (2013).
Moutsianas, L. et al. The power of gene-based rare variant methods to detect disease-associated variation and test hypotheses about complex disease. PLoS Genet. 11, e1005165 (2015).
Cardinale, C. J., Kelsen, J. R., Baldassano, R. N. & Hakonarson, H. Impact of exome sequencing in inflammatory bowel disease. World J. Gastroenterol. 19, 6721–6729 (2013).
Firestein, G. S. Evolving concepts of rheumatoid arthritis. Nature 423, 356–361 (2003).
Lee, D. M. et al. Mast cells: a cellular link between autoantibodies and inflammatory arthritis. Science 297, 1689–1692 (2002).
Pap, T., Müller-Ladner, U., Gay, R. E. & Gay, S. Fibroblast biology. Role of synovial fibroblasts in the pathogenesis of rheumatoid arthritis. Arthritis Res. 2, 361–367 (2000).
Lefèvre, S. et al. Synovial fibroblasts spread rheumatoid arthritis to unaffected joints. Nat. Med. 15, 1414–1420 (2009).
Li, P., Spolski, R., Liao, W. & Leonard, W. J. Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunol. Rev. 261, 141–156 (2014).
Lovett-Racke, A. E., Yang, Y. & Racke, M. K. Th1 versus Th17: are T cell cytokines relevant in multiple sclerosis? Biochim. Biophys. Acta 1812, 246–251 (2011).
Heng, T. S. P. et al. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Hu, X. et al. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am. J. Hum. Genet. 89, 682 (2011). This is the first study integrating GWAS loci with cell-type-specific gene expression to find pathogenic cell types of autoimmune diseases.
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2013).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Parker, S. C. J. et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl Acad. Sci. USA 110, 17921–17926 (2013).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Adrianto, I. et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 43, 253–258 (2011).
Ritchie, G. R. S., Dunham, I., Zeggini, E. & Flicek, P. Functional annotation of noncoding sequence variants. Nat. Methods 11, 294–296 (2014).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Gagliano, S. A., Barnes, M. R., Weale, M. E. & Knight, J. A. Bayesian method to incorporate hundreds of functional characteristics with association evidence to improve variant prioritization. PLoS ONE 9, e98122 (2014).
Shihab, H. A. et al. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 31, 1536–1543 (2015).
Trynka, G. et al. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am. J. Hum. Genet. 97, 139–152 (2015).
Kichaev, G. et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10, e1004722 (2014).
Gusev, A. et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 95, 535–552 (2014).
Vahedi, G. et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015). This paper describes superenhancers in T cells and reveals that superenhancers associated with RA genes are altered with the drug tofacitinib.
Stranger, B. E. et al. Population genomics of human gene expression. Nat. Genet. 39, 1217–1224 (2007).
Fairfax, B. P. et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat. Genet. 44, 502–510 (2012).
Raj, T. et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
Liang, L. et al. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 23, 716–726 (2013).
Wen, X., Luca, F. & Pique-Regi, R. Cross-population joint analysis of eQTLs: fine mapping and functional annotation. PLoS Genet. 11, e1005176 (2015).
Dimas, A. S. et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325, 1246–1250 (2009).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089 (2012).
Fu, J. et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 8, e1002431 (2012).
Gutierrez-Arcelus, M. et al. Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet. 11, e1004958 (2015).
GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Nica, A. C. et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 7, e1002003 (2011).
Hu, X. et al. Regulation of gene expression in autoimmune disease loci and the genetic basis of proliferation in CD4+ effector memory T cells. PLoS Genet. 10, e1004404 (2014).
Petretto, E. et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2, e172 (2006).
Westra, H.-J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013). This study uses two large cohorts to identify and replicate trans eQTLs in blood, highlighting an interesting example implicating SLE.
Montgomery, S. B. et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464, 773–777 (2010).
Pickrell, J. K. et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–772 (2010).
Fairfax, B. P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014).
Lee, M. N. et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343, 1246980 (2014).
Ferraro, A. et al. Interindividual variation in human T regulatory cells. Proc. Natl Acad. Sci. USA 111, E1111–E1120 (2014).
Naranbhai, V. et al. Genomic modulators of gene expression in human neutrophils. Nat. Commun. 6, 7545 (2015).
Han, J.-W. et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat. Genet. 41, 1234–1237 (2009).
Gateva, V. et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 41, 1228–1233 (2009).
Wang, S. et al. A functional haplotype of UBE2L3 confers risk for systemic lupus erythematosus. Genes Immun. 13, 380–387 (2012).
International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 40, 204–210 (2008).
Lewis, M. J. et al. UBE2L3 polymorphism amplifies NF-κB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am. J. Hum. Genet. 96, 221–234 (2015). This study describes an example of a risk locus for SLE that affects cellular phenotypes and subsequent immunophenotypes, one of which is particularly altered in patients.
Dermitzakis, E. T. Cellular genomics for complex traits. Nat. Rev. Genet. 13, 215–220 (2012).
Barreiro, L. B. et al. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc. Natl Acad. Sci. USA 109, 1204–1209 (2012).
Ye, C. J. et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science 345, 1254665 (2014). This paper, which is part of the ImmVar project, studies cell-state-specific eQTLs by subjecting CD4+ T cells to various stimuli in individuals of three different ancestral origins.
Romanoski, C. E. et al. Systems genetics analysis of gene-by-environment interactions in human cells. Am. J. Hum. Genet. 86, 399–410 (2010).
Corradin, O. et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24, 1–13 (2014).
Rakyan, V. K., Down, T. A., Balding, D. J. & Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 12, 529–541 (2011).
Rakyan, V. K. et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 7, e1002300 (2011).
Javierre, B. M. et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20, 170–179 (2010).
Gutierrez-Arcelus, M. et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2, e00523 (2013).
Kilpinen, H. et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science 342, 744–747 (2013).
McVicker, G. et al. Identification of genetic variants that affect histone modifications in human cells. Science 342, 747–749 (2013).
Banovich, N. E. et al. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 10, e1004663 (2014).
Lemire, M. et al. Long-range epigenetic regulation is conferred by genetic variation located at thousands of independent loci. Nat. Commun. 6, 6326 (2015).
Millstein, J., Zhang, B., Zhu, J. & Schadt, E. E. Disentangling molecular relationships with a causal inference test. BMC Genet. 10, 23 (2009).
Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31, 142–147 (2013).
Fullwood, M. J. et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 (2009).
Kraal, G., Weissman, I. L. & Butcher, E. C. Genetic control of T-cell subset representation in inbred mice. Immunogenetics 18, 585–592 (1983).
Amadori, A. et al. Genetic control of the CD4/CD8 T-cell ratio in humans. Nat. Med. 1, 1279–1283 (1995).
Clementi, M. et al. CD4 and CD8 T lymphocyte inheritance. Evidence for major autosomal recessive genes. Hum. Genet. 105, 337–342 (1999).
Evans, D. M., Frazer, I. H. & Martin, N. G. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 2, 250–257 (1999).
Ferreira, M. A. R. et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am. J. Hum. Genet. 86, 88–92 (2010).
Hall, M. A. et al. Genetic influence on peripheral blood T lymphocyte levels. Genes Immun. 1, 423–427 (2000).
Nalls, M. A. et al. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 7, e1002113 (2011).
Okada, Y. et al. Identification of nine novel loci associated with white blood cell subtypes in a Japanese population. PLoS Genet. 7, e1002067 (2011).
Ermann, J., Rao, D. A., Teslovich, N. C., Brenner, M. B. & Raychaudhuri, S. Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat. Rev. Rheumatol. 11, 541–551 (2015).
Maecker, H. T., McCoy, J. P. & Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 12, 191–200 (2012).
Brodin, P. et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 160, 37–47 (2015). This study measures a wide range of immunophenotypes in >200 twins, estimating the genetic and environmental contributions to each.
Orrù, V. et al. Genetic variants regulating immune cell levels in health and disease. Cell 155, 242–256 (2013). This study reports genetic associations with immune cell type abundances in >1,000 Sardinian individuals.
Roederer, M. et al. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell 161, 387–403 (2015).
Nair, R. P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 41, 199–204 (2009).
Morahan, G. et al. Association of IL12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet 360, 455–459 (2002).
International Multiple Sclerosis Genetics Consortium et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011).
Housley, W. J. et al. Genetic variants associated with autoimmunity drive NFκB signaling and responses to inflammatory stimuli. Sci. Transl Med. 7, 291ra93 (2015). This paper implicates NF-κB and its pathway in MS by showing alterations of various immunophenotypes driven by risk alleles.
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Belton, J.-M. et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012).
Buil, A. et al. Gene–gene and gene–environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet. 47, 88–91 (2015).
Sandberg, R. Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods 11, 22–24 (2014).
Pai, A. A. et al. The contribution of RNA decay quantitative trait loci to inter-individual variation in steady-state gene expression levels. PLoS Genet. 8, e1003000 (2012).
Hause, R. J. et al. Identification and validation of genetic variants that influence transcription factor and cell signaling protein levels. Am. J. Hum. Genet. 95, 194–208 (2014).
Battle, A. et al. Impact of regulatory variation from RNA to protein. Science 347, 664–667 (2015).
Wu, L. et al. Variation and genetic control of protein abundance in humans. Nature 499, 79–82 (2013).
Liu, Y. et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 11, 786 (2015).
Garge, N. et al. Identification of quantitative trait loci underlying proteome variation in human lymphoblastoid cells. Mol. Cell. Proteomics 9, 1383–1399 (2010).
Sahni, N. et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660 (2015).
Rossin, E. J. et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7, e1001273 (2011).
International Multiple Sclerosis Genetics Consortium. Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. Am. J. Hum. Genet. 92, 854–865 (2013).
Topol, E. J. Individualized medicine from prewomb to tomb. Cell 157, 241–253 (2014).
Dendrou, C. A. et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat. Genet. 41, 1011–1015 (2009).
De Jager, P. L. et al. ImmVar project: Insights and design considerations for future studies of 'healthy' immune variation. Semin. Immunol. 27, 51–57 (2015).
Klareskog, L., Rönnelid, J., Lundberg, K., Padyukov, L. & Alfredsson, L. Immunity to citrullinated proteins in rheumatoid arthritis. Annu. Rev. Immunol. 26, 651–675 (2008).
Chattopadhyay, P. K. et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat. Med. 12, 972–977 (2006).
Bendall, S. C., Nolan, G. P., Roederer, M. & Chattopadhyay, P. K. A deep profiler's guide to cytometry. Trends Immunol. 33, 323–332 (2012).
Purohit, S., Sharma, A. & She, J. X. Luminex and other multiplex high throughput technologies for the identification of, and host response to, environmental triggers of type 1 diabetes. Biomed. Res. Int. 2015, 326918 (2015).
Kondrat, R. W., McClusky, G. A. & Cooks, R. G. Multiple reaction monitoring in mass spectrometry/mass spectrometry for direct analysis of complex mixtures. Anal. Chem. 50, 2017–2021 (1978).
Gillet, L. C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012).
Bowcock, A. M. & Krueger, J. G. Getting under the skin: the immunogenetics of psoriasis. Nat. Rev. Immunol. 5, 699–711 (2005).
Selmi, C., Lu, Q. & Humble, M. C. Heritability versus the role of the environment in autoimmunity. J. Autoimmun. 39, 249–252 (2012).
Akkoc, N. & Khan, M. A. Overestimation of the prevalence of ankylosing spondylitis in the Berlin study: comment on the article by Braun et al. Arthritis Rheum. 52, 4048–4049 (2005).
Ng, S. C. et al. Epidemiology of spondyloarthritis in the People's Republic of China: review of the literature and commentary. Semin. Arthritis Rheum. 37, 39–47 (2007).
Liu, J. Z. et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 44, 1137–1141 (2012).
Villanueva, R., Greenberg, D. A., Davies, T. F. & Tomer, Y. Sibling recurrence risk in autoimmune thyroid disease. Thyroid 13, 761–764 (2003).
Brix, T. H., Kyvik, K. O., Christensen, K. & Hegedüs, L. Evidence for a major role of heredity in Graves' disease: a population-based study of two Danish twin cohorts. J. Clin. Endocrinol. Metab. 86, 930–934 (2001).
Prahalad, S. et al. Quantification of the familial contribution to juvenile idiopathic arthritis. Arthritis Rheum. 62, 2525–2529 (2010).
Sadovnick, A. D. et al. A population-based study of multiple sclerosis in twins: update. Ann. Neurol. 33, 281–285 (1993).
Chen, G.-B. et al. Estimation and partitioning of (co)heritability of inflammatory bowel disease from GWAS and immunochip data. Hum. Mol. Genet. 23, 4710–4720 (2014).
Jones, D. E., Watt, F. E., Metcalf, J. V, Bassendine, M. F. & James, O. F. Familial primary biliary cirrhosis reassessed: a geographically-based population study. J. Hepatol. 30, 402–407 (1999).
Acknowledgements
This work is supported in part by funding from the Swiss National Science Foundation (M.G.-A.), the US National Institutes of Health (1R01AR063759, 5U01GM092691-05, 1UH2AR067677-01 and U19 AI111224-01 (S.R.)) and the Doris Duke Charitable Foundation Grant #2013097 (S.R.). The authors thank the Raychaudhuri laboratory for their feedback, in particular H.-J. Westra and C. M. Hong for their careful reading of the manuscript and C. Y. Fonseka for figure support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Adaptive immune system
-
The part of the immune system that can react to specific pathogens and remember them to elicit a faster immune response if they are re-encountered.
- Autoimmunity
-
When the immune system elicits an immune response against its own body (or non-pathogenic antigens that are commonly found in the body, such as commensal bacteria).
- Type 1 diabetes mellitus
-
(T1DM). A disease characterized by autoreactivity to insulin-secreting β-cells found in the pancreas, resulting in high glucose levels in blood and urine.
- Inflammatory bowel disease
-
(IBD). A term that groups together several conditions, such as Crohn's disease and ulcerative colitis, that are characterized by inflammation of the small intestine and colon. The inflammation is thought to be caused by immune reaction to commensal bacteria.
- Rheumatoid arthritis
-
(RA). Autoimmune disease causing joint inflammation and destruction of cartilage and bone. Patients with RA can present autoantibodies (seropositive) or not (seronegative).
- Systemic lupus erythematosus
-
(SLE). A disease characterized by autoreactivity to ubiquitous molecules, such as DNA or chromatin proteins, which can cause immune reactions against many parts of the body, some of the most common being joints, skin, blood vessels, kidneys and nervous system.
- Multiple sclerosis
-
(MS). A condition of autoimmunity against the myelin sheath that protects the central nervous system.
- Self-tolerance
-
The mechanism by which the immune system does not attack its own body.
- Genome-wide association studies
-
(GWAS). Studies in which genetic variants are ascertained across the whole genome in thousands of patients and thousands of controls, and statistical genetic associations (but not causality) with disease are tested.
- Effect sizes
-
Quantitative measures of the strength of events. In genetic association studies, the odds ratio is often used as an effect size measure, as it reflects how many times more likely it is to find the susceptibility variant in cases versus controls.
- Immunophenotypes
-
In this article, we define immunophenotypes as quantitative traits that reflect immune function. They include cell-type abundances, cell proliferation, signalling response, and cytokine and chemokine serum protein levels.
- Cytokine
-
A type of small cell-signalling protein that is secreted by some cells and affects the behaviour of others; cytokines are crucial participants in the immune response.
- Psoriasis
-
An autoimmune disease affecting the skin.
- Crohn's disease
-
Inflammatory bowel disease presenting autoreactivity in any part of the gastrointestinal tract.
- Ulcerative colitis
-
Inflammatory bowel disease mainly affecting the colon.
- Coeliac disease
-
An autoimmune disease of the small intestine caused by an immune reaction to wheat.
- Linkage analysis
-
A genetic association method that uses linkage disequilibrium and recombination principles in families to find markers linked to a disease locus (markers co-segregating with the disease phenotype).
- Human leukocyte antigen
-
(HLA). A type of surface protein that presents antigens. HLA class I proteins present antigens from inside the cell and are expressed by most cells, whereas HLA class II proteins are expressed only by antigen-presenting cells and present extracellular antigens to T cells.
- Nuclear factor-κB
-
(NF-κB). An important transcription factor involved in the immune response and B cell development.
- Candidate gene studies
-
A genetic association method that looks for alleles of a candidate gene (for example, based on a priori functional knowledge) that are associated with a disease, typically using a case-control study design.
- Linkage disequilibrium
-
(LD). A phenomenon describing the nonrandom association (or co-segregation) between two alleles of different loci.
- Trans-ancestral cohorts
-
Groups of individuals pertaining to different ancestry populations.
- Imputation
-
Inference of alleles of non-genotyped variants based on nearby observed genotypes and using a reference panel from which the correlation structure between variants (or haplotypes) can be learnt.
- Missing heritability
-
A concept referring to the heritability that has not been explained by known disease susceptibility loci (despite so many having been discovered with genome-wide association studies).
- Exome sequencing
-
A technique that captures the protein-coding sequences of the genome (exons) and sequences them.
- Enhancers
-
Distant transcriptional regulatory elements, often regulating expression in a cell-type- and cell-state-specific manner in cis, and often marked by specific chromatin modifications.
- Super enhancer
-
Several enhancer marks clustered together; super enhancers tend to be more cell-type specific than typical enhancer marks.
- Expression quantitative trait loci
-
(eQTLs). Refers to a variant whose genotypes correlate with gene expression levels.
- QTLs
-
(Quantitative trait loci). Loci in the genome that are associated with a quantitative trait. For example, a single-nucleotide polymorphism associated with gene expression levels can be called an expression QTL (eQTL).
- Mendelian randomization
-
A method for inferring causality of a trait (or modifiable exposure) on disease, taking advantage of the randomized trial nature of genetic variation (assuming a random mating pattern).
- Allele-specific expression
-
A phenomenon that occurs when one of the two alleles of a gene in an individual is expressed more than the other.
Rights and permissions
About this article
Cite this article
Gutierrez-Arcelus, M., Rich, S. & Raychaudhuri, S. Autoimmune diseases — connecting risk alleles with molecular traits of the immune system. Nat Rev Genet 17, 160–174 (2016). https://doi.org/10.1038/nrg.2015.33
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg.2015.33
This article is cited by
-
A novel missense mutation in the AIRE gene underlying autoimmune polyglandular syndrome type 1
Immunogenetics (2024)
-
An autoimmune pleiotropic SNP modulates IRF5 alternative promoter usage through ZBTB3-mediated chromatin looping
Nature Communications (2023)
-
Why are you hitting yourself? Whole-exome sequencing diagnosis of monogenic autoimmunity
Journal of Genetics (2023)
-
A common human MLKL polymorphism confers resistance to negative regulation by phosphorylation
Nature Communications (2023)
-
Germline genetic variation and predicting immune checkpoint inhibitor induced toxicity
npj Genomic Medicine (2022)