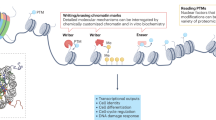
Key Points
-
Synthetic approaches are increasingly being applied to study and harness chromatin biology.
-
Residue-specific histone modifications and site-specific chromatin modifications provide powerful approaches to address key questions about the 'histone code' and chromatin-based gene expression 'memory'.
-
The synthetic positioning of nucleosomes enables fine-tuning of gene expression and can drive complex nonlinear gene expression programmes.
-
Chromatin exhibits spatial spreading of histone marks and gene regulation. The boundaries of these spatial regions can be synthetically controlled using both protein and DNA sequence elements.
-
Long-range chromatin interactions have been synthetically recapitulated, providing insights into their roles in gene regulation and development, as well as potential approaches to control gene expression.
-
Continued collaboration between chromatin biology and engineering will reveal mechanisms underlying gene regulation and the computational potential of chromatin. It will also provide a quantitative framework to harness chromatin in cellular engineering applications.
Abstract
As synthetic biology approaches are extended to diverse applications throughout medicine, biotechnology and basic biological research, there is an increasing need to engineer yeast, plant and mammalian cells. Eukaryotic genomes are regulated by the diverse biochemical and biophysical states of chromatin, which brings distinct challenges, as well as opportunities, over applications in bacteria. Recent synthetic approaches, including 'epigenome editing', have allowed the direct and functional dissection of many aspects of physiological chromatin regulation. These studies lay the foundation for biomedical and biotechnological engineering applications that could take advantage of the unique combinatorial and spatiotemporal layers of chromatin regulation to create synthetic systems of unprecedented sophistication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Kotula, J. W. et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl Acad. Sci. USA 111, 4838–4843 (2014).
Ye, H., Aubel, D. & Fussenegger, M. Synthetic mammalian gene circuits for biomedical applications. Curr. Opin. Chem. Biol. 17, 910–917 (2013).
Struhl, K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98, 1–4 (1999).
Ellis, L., Atadja, P. W. & Johnstone, R. W. Epigenetics in cancer: targeting chromatin modifications. Mol. Cancer Ther. 8, 1409–1420 (2009).
Ernst, J. et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011).
Gaspar-Maia, A., Alajem, A., Meshorer, E. & Ramalho-Santos, M. Open chromatin in pluripotency and reprogramming. Nature Rev. Mol. Cell Biol. 12, 36–47 (2011).
Onder, T. T. et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598–602 (2012).
Rheinbay, E., Louis, David, N., Bernstein, Bradley, E. & Suvà, Mario, L. A. Tell-tail sign of chromatin: histone mutations drive pediatric glioblastoma. Cancer Cell 21, 329–331 (2012).
Schuster-Böckler, B. & Lehner, B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488, 504–507 (2012).
Zhu, J. et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642–654 (2013).
Jost, J. & Saluz, H. DNA Methylation: Molecular Biology and Biological Significance (Birkhäuser Basel, 2011).
Ballare, C. et al. Nucleosome-driven transcription factor binding and gene regulation. Mol. Cell 49, 67–79 (2013).
Kornberg, R. D. & Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999).
Narlikar, G. J., Fan, H. Y. & Kingston, R. E. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487 (2002).
Benveniste, D., Sonntag, H. J., Sanguinetti, G. & Sproul, D. Transcription factor binding predicts histone modifications in human cell lines. Proc. Natl Acad. Sci. USA 111, 13367–13372 (2014).
Lanctot, C., Cheutin, T., Cremer, M., Cavalli, G. & Cremer, T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev. Genet. 8, 104–115 (2007).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Ram, O. et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147, 1628–1639 (2011).
Zhou, V. W., Goren, A. & Bernstein, B. E. Charting histone modifications and the functional organization of mammalian genomes. Nature Rev. Genet. 12, 7–18 (2011).
Clapier, C. R. & Cairns, B. R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009).
Allfrey, V. G., Faulkner, R. & Mirsky, A. E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA 51, 786–794 (1964).
Vidali, G., Gershey, E. L. & Allfrey, V. G. Chemical studies of histone acetylation. The distribution of ε-N-acetyllysine in calf thymus histones. J. Biol. Chem. 243, 6361–6366 (1968).
Tan, M. et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Henikoff, S. & Shilatifard, A. Histone modification: cause or cog? Trends Genet. 27, 389–396 (2011).
Turner, B. M. The adjustable nucleosome: an epigenetic signaling module. Trends Genet. 28, 436–444 (2012).
Pick, H., Kilic, S. & Fierz, B. Engineering chromatin states: chemical and synthetic biology approaches to investigate histone modification function. Biochim. Biophys. Acta 1839, 644–656 (2014).
He, S. et al. Facile synthesis of site-specifically acetylated and methylated histone proteins: reagents for evaluation of the histone code hypothesis. Proc. Natl Acad. Sci. USA 100, 12033–12038 (2003).
Shimko, J. C., Howard, C. J., Poirier, M. G. & Ottesen, J. J. Preparing semisynthetic and fully synthetic histones H3 and H4 to modify the nucleosome core. Methods Mol. Biol. 981, 177–192 (2013).
Nguyen, U. T. et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nature Methods 11, 834–840 (2014). By reconstituting chemically synthesized histones with barcoded DNA molecules, this study was able to pool diverse histone–DNA complexes, thus greatly reducing the number of biochemical assays needed when exploring a large combinatorial histone modification space.
Neumann, H. et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3K56 acetylation. Mol. Cell 36, 153–163 (2009).
Davis, L. & Chin, J. W. Designer proteins: applications of genetic code expansion in cell biology. Nature Rev. Mol. Cell Biol. 13, 168–182 (2012).
Nguyen, D. P., Garcia Alai, M. M., Kapadnis, P. B., Neumann, H. & Chin, J. W. Genetically encoding Nε-methyl-l-lysine in recombinant histones. J. Am. Chem. Soc. 131, 14194–14195 (2009).
Nguyen, D. P., Garcia Alai, M. M., Virdee, S. & Chin, J. W. Genetically directing ε-N, N-dimethyl-l-lysine in recombinant histones. Chem. Biol. 17, 1072–1076 (2010).
Chin, J. W. et al. An expanded eukaryotic genetic code. Science 301, 964–967 (2003).
Dion, M. F., Altschuler, S. J., Wu, L. F. & Rando, O. J. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl Acad. Sci. USA 102, 5501–5506 (2005).
Kim, J. A., Hsu, J. Y., Smith, M. M. & Allis, C. D. Mutagenesis of pairwise combinations of histone amino-terminal tails reveals functional redundancy in budding yeast. Proc. Natl Acad. Sci. USA 109, 5779–5784 (2012).
Dai, J. et al. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134, 1066–1078 (2008).
Siddique, A. N. et al. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3A–Dnmt3L single-chain fusion protein with increased DNA methylation activity. J. Mol. Biol. 425, 479–491 (2013).
Brent, R. & Ptashne, M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43, 729–736 (1985).
de Groote, M. L., Verschure, P. J. & Rots, M. G. Epigenetic editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 40, 10596–10613 (2012).
Gaj, T., Gersbach, C. A. & Barbas, C. F. 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 (2013).
Maeder, M. L. et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE–TET1 fusion proteins. Nature Biotech. 31, 1137–1142 (2013).
Chen, H. et al. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res. 42, 1563–1574 (2014).
Carvin, C. D., Parr, R. D. & Kladde, M. P. Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Res. 31, 6493–6501 (2003).
Rivenbark, A. G. et al. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 7, 350–360 (2012).
Nunna, S., Reinhardt, R., Ragozin, S. & Jeltsch, A. Targeted methylation of the epithelial cell adhesion molecule (EpCAM) promoter to silence its expression in ovarian cancer cells. PLoS ONE 9, e87703 (2014).
Falahi, F. et al. Towards sustained silencing of HER2/neu in cancer by epigenetic editing. Mol. Cancer Res. 11, 1029–1039 (2013). References 48–50 show the potential advantage, over other gene repression technologies, of site-specific DNA methyltransferases in stably inhibiting oncogenes.
Snowden, A. W., Gregory, P. D., Case, C. C. & Pabo, C. O. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol. 12, 2159–2166 (2002). The combined use of site-specific recruitment, minimal histone-modifying catalytic domains and catalytically dead mutants in this study provides strong evidence for the causative repressive nature of H3K9 methylation.
Mendenhall, E. M. et al. Locus-specific editing of histone modifications at endogenous enhancers. Nature Biotech. 31, 1133–1136 (2013). In this study, TALEs are used to alter the epigenetic state of endogenous enhancers, which enables the functional identification of their target genes in a highly native and physiological context.
Keung, A. J., Bashor, C. J., Kiriakov, S., Collins, J. J. & Khalil, A. S. Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell 158, 110–120 (2014). A library of more than 200 chromatin-modifying proteins were fused to ZFs and screened against diverse synthetic reporter architectures to reveal chromatin-based combinatorial and spatiotemporal regulatory behaviours.
Meister, G. E., Chandrasegaran, S. & Ostermeier, M. Heterodimeric DNA methyltransferases as a platform for creating designer zinc finger methyltransferases for targeted DNA methylation in cells. Nucleic Acids Res. 38, 1749–1759 (2010).
Chaikind, B., Kilambi, K. P., Gray, J. J. & Ostermeier, M. Targeted DNA methylation using an artificially bisected M.HhaI fused to zinc fingers. PLoS ONE 7, e44852 (2012).
Nomura, W. & Barbas, C. F. 3rd . In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J. Am. Chem. Soc. 129, 8676–8677 (2007).
Maeder, M. L. et al. CRISPR RNA-guided activation of endogenous human genes. Nature Methods 10, 977–979 (2013).
Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR–Cas9-based transcription factors. Nature Methods 10, 973–976 (2013).
Moazed, D. Mechanisms for the inheritance of chromatin states. Cell 146, 510–518 (2011).
Ptashne, M. Epigenetics: core misconcept. Proc. Natl Acad. Sci. USA 110, 7101–7103 (2013).
Andrulis, E. D., Neiman, A. M., Zappulla, D. C. & Sternglanz, R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394, 592–595 (1998).
Triolo, T. & Sternglanz, R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381, 251–253 (1996).
Chien, C. T., Buck, S., Sternglanz, R. & Shore, D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75, 531–541 (1993).
Buck, S. W. & Shore, D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9, 370–384 (1995).
Lustig, A. J., Liu, C., Zhang, C. & Hanish, J. P. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2483–2495 (1996).
Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012). This paper describes an experimental system to dynamically recruit HP1 to specific genomic sites, which allowed measurement and modelling of the kinetics of heterochromatin establishment and memory.
Laurent, B. C., Treitel, M. A. & Carlson, M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol. Cell. Biol. 10, 5616–5625 (1990).
Li, F. et al. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 35, 100–112 (2007).
Smith, A. E., Hurd, P. J., Bannister, A. J., Kouzarides, T. & Ford, K. G. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J. Biol. Chem. 283, 9878–9885 (2008).
Ragunathan, K., Jih, G. & Moazed, D. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science http://dx.doi.org/10.1126/science.1258699 (2014). Using transient site-specific induction of ectopic H3K9 methylation, this study shows that histone modification can itself act epigenetically and be inherited in the absence of sequence-specific factors, DNA methylation or RNAi.
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Grimmer, M. R. et al. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res. 42, 10856–10868 (2014).
Kuscu, C., Arslan, S., Singh, R., Thorpe, J. & Adli, M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nature Biotech. 32, 677–683 (2014).
Smith, C. et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15, 12–13 (2014).
Wu, X., Kriz, A. J. & Sharp, P. A. Target specificity of the CRISPR–Cas9 system. Quantitative Biol. 2, 59–70 (2014).
Gabriel, R. et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature Biotech. 29, 816–823 (2011).
Guilinger, J. P. et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nature Methods 11, 429–435 (2014).
Pattanayak, V., Ramirez, C. L., Joung, J. K. & Liu, D. R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nature Methods 8, 765–770 (2011).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases. Nature Biotech. http://dx.doi.org/10.1038/nbt.3117 (2014).
Guilinger, J. P., Thompson, D. B. & Liu, D. R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature Biotech. 32, 577–582 (2014).
Tsai, S. Q. et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature Biotech. 32, 569–576 (2014).
Khalil, A. S. et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell 150, 647–658 (2012).
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M. & Joung, J. K. Improving CRISPR–Cas nuclease specificity using truncated guide RNAs. Nature Biotech. 32, 279–284 (2014).
Tan, S. et al. Zinc-finger protein-targeted gene regulation: genomewide single-gene specificity. Proc. Natl Acad. Sci. USA 100, 11997–12002 (2003).
Haynes, K. A. & Silver, P. A. Synthetic reversal of epigenetic silencing. J. Biol. Chem. 286, 27176–27182 (2011).
Fierz, B. & Muir, T. W. Chromatin as an expansive canvas for chemical biology. Nature Chem. Biol. 8, 417–427 (2012).
Ruthenburg, A. J., Li, H., Patel, D. J. & Allis, C. D. Multivalent engagement of chromatin modifications by linked binding modules. Nature Rev. Mol. Cell Biol. 8, 983–994 (2007).
Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013). This paper describes light-activated induction of site-specific transcriptional activation and histone modifications.
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Knezetic, J. A. & Luse, D. S. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell 45, 95–104 (1986).
Lorch, Y., LaPointe, J. W. & Kornberg, R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49, 203–210 (1987).
Han, M. & Grunstein, M. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55, 1137–1145 (1988).
Hughes, A. L., Jin, Y., Rando, O. J. & Struhl, K. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol. Cell 48, 5–15 (2012).
Shrader, T. E. & Crothers, D. M. Artificial nucleosome positioning sequences. Proc. Natl Acad. Sci. USA 86, 7418–7422 (1989).
Tanaka, S., Zatchej, M. & Thoma, F. Artificial nucleosome positioning sequences tested in yeast minichromosomes: a strong rotational setting is not sufficient to position nucleosomes in vivo. EMBO J. 11, 1187–1193 (1992).
Raveh-Sadka, T. et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nature Genet. 44, 743–750 (2012). This study generates a library of synthetic promoters with fine-tuned activity by altering the number and location of nucleosome-disfavouring sequences.
Lam, F. H., Steger, D. J. & O'Shea, E. K. Chromatin decouples promoter threshold from dynamic range. Nature 453, 246–250 (2008).
Mirny, L. A. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl Acad. Sci. USA 107, 22534–22539 (2010).
Bi, X. & Broach, J. R. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 13, 1089–1101 (1999).
Bi, X., Yu, Q., Sandmeier, J. J. & Zou, Y. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 24, 2118–2131 (2004).
Donze, D. & Kamakaka, R. T. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20, 520–531 (2001).
Ishii, K., Arib, G., Lin, C., Van Houwe, G. & Laemmli, U. K. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109, 551–562 (2002).
Marquardt, S. et al. A chromatin-based mechanism for limiting divergent noncoding transcription. Cell 157, 1712–1723 (2014). By integrating a bidirectional fluorescent protein reporter into each strain of the non-essential yeast deletion library, the authors identified the CAF-I complex as an important regulator of divergent transcription.
Cremer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Rev. Genet. 2, 292–301 (2001).
de Bruin, D., Zaman, Z., Liberatore, R. A. & Ptashne, M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409, 109–113 (2001).
Zaman, Z., Heid, C. & Ptashne, M. Telomere looping permits repression “at a distance” in yeast. Curr. Biol. 12, 930–933 (2002).
Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012).
Deng, W. et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158, 849–860 (2014). References 107 and 108 describe how the synthetically induced looping of chromatin between the promoters and LCRs of the β- and γ-globin loci can activate these genes.
Brennan, T. A. & Wilson, J. M. The special case of gene therapy pricing. Nature Biotech. 32, 874–876 (2014).
Noordermeer, D. et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nature Cell Biol. 13, 944–951 (2011).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Fanucchi, S., Shibayama, Y., Burd, S., Weinberg, M. S. & Mhlanga, M. M. Chromosomal contact permits transcription between coregulated genes. Cell 155, 606–620 (2013).
Clowney, E. J. et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151, 724–737 (2012).
Lacefield, S., Lau, D. T. & Murray, A. W. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nature Cell Biol. 11, 1116–1120 (2009).
Kagansky, A. et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324, 1716–1719 (2009).
Nannas, N. J. & Murray, A. W. Tethering sister centromeres to each other suggests the spindle checkpoint detects stretch within the kinetochore. PLoS Genet. 10, e1004492 (2014).
Reddy, K. L., Zullo, J. M., Bertolino, E. & Singh, H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247 (2008).
Kind, J. et al. Single-cell dynamics of genome–nuclear lamina interactions. Cell 153, 178–192 (2013).
Aragon-Alcaide, L. & Strunnikov, A. V. Functional dissection of in vivo interchromosome association in Saccharomyces cerevisiae. Nature Cell Biol. 2, 812–818 (2000).
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013).
Alon, U., Surette, M. G., Barkai, N. & Leibler, S. Robustness in bacterial chemotaxis. Nature 397, 168–171 (1999).
Barkai, N. & Leibler, S. Robustness in simple biochemical networks. Nature 387, 913–917 (1997).
Dymond, J. S. et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 477, 471–476 (2011).
Sander, J. D. & Joung, J. K. CRISPR–Cas systems for editing, regulating and targeting genomes. Nature Biotech. 32, 347–355 (2014).
Tischer, D. & Weiner, O. D. Illuminating cell signalling with optogenetic tools. Nature Rev. Mol. Cell Biol. 15, 551–558 (2014).
Suva, M. L., Riggi, N. & Bernstein, B. E. Epigenetic reprogramming in cancer. Science 339, 1567–1570 (2013).
Meissner, A. Epigenetic modifications in pluripotent and differentiated cells. Nature Biotech. 28, 1079–1088 (2010).
Buhler, M., Verdel, A. & Moazed, D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125, 873–886 (2006).
Jiang, J. et al. Translating dosage compensation to trisomy 21. Nature 500, 296–300 (2013). By inserting the long non-coding RNA XIST into one of three copies of chromosome 21 in Down syndrome pluripotent stem cells, this study was able to completely silence the extra chromosome, suggesting a potential therapeutic avenue for many polyploidy diseases.
Keller, C., Kulasegaran-Shylini, R., Shimada, Y., Hotz, H. R. & Buhler, M. Noncoding RNAs prevent spreading of a repressive histone mark. Nature Struct. Mol. Biol. 20, 1340 (2013).
Acknowledgements
This work was supported by a US National Institutes of Health (NIH)-National Institute of General Medical Sciences (NIGMS) Ruth L. Kirschstein Postdoctoral Fellowship (A.J.K.), an NIH Director's Pioneer Award (J.K.J.), a Defense Advanced Research Projects Agency grant (J.K.J., A.S.K., and J.J.C.), start-up funds from the Department of Biomedical Engineering at Boston University (A.S.K.), a National Science Foundation CAREER Award (A.S.K.), a NIH R24 (J.J.C.), the Wyss Institute for Biologically Inspired Engineering (J.J.C.) and the Howard Hughes Medical Institute (J.J.C.). Owing to space limitations, the authors regret that many important publications were not able to be discussed in this Review.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. J.K.J.'s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. A.J.K., A.S.K. and J.J.C. declare no competing interests.
Glossary
- Nucleosomes
-
Octamer protein complexes in which an octamer is comprised of two copies each of H3, H4, H2A and H2B histone proteins.
- Non-coding RNAs
-
Functional RNA molecules that are not translated into proteins.
- Lysine acetylation
-
A post-translational modification in which an acetyl group reacts with the primary amine on the side chain of a lysine residue.
- Correlation
-
A relationship between two or more variables. The correlation between the occupancy of a chromatin modification and transcriptional activity does not directly prove that the modification causes transcriptional activity, or vice versa.
- Pleiotropy
-
The phenomenon whereby one gene influences multiple other seemingly unrelated genes or traits.
- Unnatural amino acid
-
An amino acid that is not naturally encoded or found in the genetic code of an organism.
- Chemical ligation
-
A chemical reaction that links a fully chemically derived peptide to the end of a recombinant protein.
- Zinc-finger
-
(ZF). A small protein structural motif coordinated to one or more zinc ions that stabilize its fold.
- Transcription activator-like effector
-
(TALE). A bacterial protein with a variable number of 34-amino-acid repeats, of which 2 residues specify binding to a DNA base.
- CRISPR
-
(Clustered regularly interspaced short palindromic repeat). An important part of a prokaryotic adaptive immune system that uses short RNAs to guide the CRISPR-associated 9 (Cas9) nuclease to specific targets, which cleaves foreign DNA elements such as plasmids and phage genomes.
- RNA interference
-
(RNAi). A biological process that inhibits gene expression through RNA molecules interacting and interfering with specific mRNA molecules.
- FKBP–FRB
-
A protein pair consisting of FKBP and FRB, which dimerize by mutually binding to the small molecule rapamycin.
- PYL–ABI
-
A protein pair consisting of PYL and ABI, which dimerize by mutually binding to abscisic acid.
- Avidity
-
The accumulated strength of multiple affinities from multivalent non-covalent binding interactions.
- Cry2 and CIB1
-
(Cryptochrome 2 and cryptochrome-interacting basic helix–loop–helix 1). A pair of proteins that dimerize at the subsecond timescale upon blue-light exposure and that dissociate on the minute timescale.
- Nuclear lamina
-
A dense fibrillar network of intermediate filament proteins at the periphery of the nucleus.
- Telomeres
-
The regions at the ends of chromosomes comprised of repetitive nucleotide sequences that are typically repressed by heterochromatin.
- Auxotrophic markers
-
Genes absent in an organism that normally produce organic compounds required for survival of the organism.
- Yeast knockout library
-
A collection of yeast strains, each of which harbour a knockout allele for a single gene. Strains are either haploid and have a non-essential gene knocked out, or diploid and have the knockout allele in a heterozygous state.
- β-globin
-
A subunit of the major haemoglobin complex found in adult mammals.
- Locus control region
-
(LCR). A genomic region that enhances the expression of genes from a distance.
- Kinetochores
-
The protein structures assembled on the centromere to which spindle fibres attach during cell division to pull sister chromatids apart.
- Centromeres
-
The genetic loci on chromosomes that link sister chromatids during mitosis and on which kinetochores assemble.
Rights and permissions
About this article
Cite this article
Keung, A., Joung, J., Khalil, A. et al. Chromatin regulation at the frontier of synthetic biology. Nat Rev Genet 16, 159–171 (2015). https://doi.org/10.1038/nrg3900
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3900
This article is cited by
-
Upstream Activation Sequence Can Function as an Insulator for Chromosomal Regulation of Heterologous Pathways Against Position Effects in Saccharomyces cerevisiae
Applied Biochemistry and Biotechnology (2022)
-
CRISPR technologies for precise epigenome editing
Nature Cell Biology (2021)
-
Engineering a growth-phase-dependent biosynthetic pathway for carotenoid production in Saccharomyces cerevisiae
Journal of Industrial Microbiology and Biotechnology (2020)
-
Digital logic circuits in yeast with CRISPR-dCas9 NOR gates
Nature Communications (2017)
-
Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer
Journal of Experimental & Clinical Cancer Research (2016)