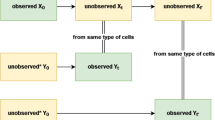
Key Points
-
Signalling networks convert observations about the environment into chemical and physical representations that ultimately modulate cellular behaviour.
-
Signalling networks are comprised of sensors, transducers and actuators.
-
The more transducers there are between a sensor and its cognate actuator, the greater the opportunity for a signal to be influenced by other signals.
-
Qualitative modelling approaches provide insights into how a signal is propagated through or integrated into a network.
-
Signalling-network reconstructions represent our knowledge regarding the network in a format that is amenable to conversion into mathematical models.
-
Signalling networks are comprised of components that are expressed in temporal fashion and may be distributed heterogeneously.
-
Signalling networks evolve to aid the organism in which they are located to overcome its specific challenge. Therefore, despite a high degree of similarity of parts, related organisms may have substantially different signalling networks.
-
The symbolic nature of signals makes it easier for signalling networks to be rewired by evolution than is the case for metabolic networks.
Abstract
Biological signalling networks allow living organisms to issue an integrated response to current conditions and make limited predictions about future environmental changes. Small-scale dynamic models of signalling cascades, including mitogen-activated protein kinase cascades, have been developed to generate hypotheses about signal transduction. Owing to technical limitations, these models and the hypotheses they generate have focused on a limited subset of signalling molecules. Now that we can simultaneously measure a substantial portion of the molecular components of a cell, we can begin to develop and test systems-level models of cellular signalling and regulatory processes, therefore gaining insights into the 'thought' processes of a cell.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Hyduke, D. R., Amundson, S. A. & Fornace, A. J. Jr. in Handbook of Cell Signaling 2nd edn Vol. 3 (eds Bradshaw, R. A. & Dennis, E. A.) 2107–2125 (Academic Press, 2009).
Tovar, C. et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl Acad. Sci. USA 103, 1888–1893 (2006).
Bulavin, D. V. et al. Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16Ink4a-p19Arf pathway. Nature Genet. 36, 343–350 (2004).
Lavelle, C., Salles, B. & Wiesmuller, L. DNA repair, damage signaling and carcinogenesis. DNA Repair (Amst.) 7, 670–680 (2008).
Abbott, D. W., Wilkins, A., Asara, J. M. & Cantley, L. C. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 14, 2217–2227 (2004).
Franke, R. et al. Host–pathogen systems biology: logical modelling of hepatocyte growth factor and Helicobacter pylori induced c-Met signal transduction. BMC Syst. Biol. 2, 4 (2008).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Wang, B., Xiao, Z. & Ren, E. C. Redefining the p53 response element. Proc. Natl Acad. Sci. USA 106, 14373–14378 (2009).
Huang, S. S. & Fraenkel, E. Integrating proteomic, transcriptional, and interactome data reveals hidden components of signaling and regulatory networks. Sci. Signal. 2, ra40 (2009).
Carthew, R. W. & Sontheimer, E. J. Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009).
Figueroa-Bossi, N., Valentini, M., Malleret, L. & Bossi, L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 23, 2004–2015 (2009).
Bhalla, U. S. & Iyengar, R. Emergent properties of networks of biological signaling pathways. Science 283, 381–387 (1999).
Li, F., Thiele, I., Jamshidi, N. & Palsson, B. Ø. Identification of potential pathway mediation targets in Toll-like receptor signaling. PLoS Comput. Biol. 5, e1000292 (2009). This paper presents one of the largest signalling-network reconstructions to date. The authors used the stoichiometric approach to identify novel signalling pathways in T cells.
Duarte, N. C. et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA 104, 1777–1782 (2007).
Feist, A. M. et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1,260 ORFs and thermodynamic information. Mol. Syst. Biol. 3, 121 (2007).
Herrgard, M. J. et al. A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nature Biotech. 26, 1155–1160 (2008).
Raghunathan, A., Reed, J., Shin, S., Palsson, B. & Daefler, S. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host–pathogen interaction. BMC Syst. Biol. 3, 38 (2009).
Vo, T. D., Greenberg, H. J. & Palsson, B. Ø. Reconstruction and functional characterization of the human mitochondrial metabolic network based on proteomic and biochemical data. J. Biol. Chem. 279, 39532–39540 (2004).
Bouwmeester, T. et al. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nature Cell Biol. 6, 97–105 (2004).
Carter, G. W. et al. Prediction of phenotype and gene expression for combinations of mutations. Mol. Syst. Biol. 3, 96 (2007).
Pawson, T. Protein modules and signalling networks. Nature 373, 573–580 (1995).
Lee, I. et al. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nature Genet. 40, 181–188 (2008).
Bhalla, U. S. Understanding complex signaling networks through models and metaphors. Prog. Biophys. Mol. Biol. 81, 45–65 (2003).
Singh, A. H., Wolf, D. M., Wang, P. & Arkin, A. P. Modularity of stress response evolution. Proc. Natl Acad. Sci. USA 105, 7500–7505 (2008). This study introduces engineering ontology for the classification of signalling pathway elements and explores the evolution of modularity in bacterial and archaeal stress responses.
Sivakumaran, S., Hariharaputran, S., Mishra, J. & Bhalla, U. S. The Database of Quantitative Cellular Signaling: management and analysis of chemical kinetic models of signaling networks. Bioinformatics 19, 408–415 (2003). This paper describes the establishment of the Database of Quantitative Cellular Signaling. This is a repository of functional kinetic models of signalling pathways that serves as a useful introduction to the kinetic modelling of signalling.
Kohn, K. W. & Aladjem, M. I. Circuit diagrams for biological networks. Mol. Syst. Biol. 2, 2006.0002 (2006).
Friedman, A. & Perrimon, N. Genetic screening for signal transduction in the era of network biology. Cell 128, 225–231 (2007). A thought-provoking essay that illustrates the demand for new conceptual frameworks to deal with signalling networks.
Barabasi, A. L. & Oltvai, Z. N. Network biology: understanding the cell's functional organization. Nature Rev. Genet. 5, 101–113 (2004). A Review that explains a variety of abstract features of intracellular organization, including modularity.
Feinerman, O., Veiga, J., Dorfman, J. R., Germain, R. N. & Altan-Bonnet, G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science 321, 1081–1084 (2008).
Gama-Castro, S. et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 36, D120–D124 (2008).
Oda, K. & Kitano, H. A comprehensive map of the Toll-like receptor signaling network. Mol. Syst. Biol. 2, 2006.0015 (2006).
Feist, A. M., Herrgard, M. J., Thiele, I., Reed, J. L. & Palsson, B. Ø. Reconstruction of biochemical networks in microorganisms. Nature Rev. Microbiol. 7, 129–143 (2009).
Papin, J. A., Hunter, T., Palsson, B. Ø. & Subramaniam, S. Reconstruction of cellular signalling networks and analysis of their properties. Nature Rev. Mol. Cell Biol. 6, 99–111 (2005).
Famili, I., Forster, J., Nielsen, J. & Palsson, B. Ø. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl Acad. Sci. USA 100, 13134–13139 (2003).
Feist, A. M. & Palsson, B. Ø. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nature Biotech. 26, 659–667 (2008).
Suthers, P. F., Zomorrodi, A. & Maranas, C. D. Genome-scale gene/reaction essentiality and synthetic lethality analysis. Mol. Syst. Biol. 5, 301 (2009).
Feist, A. M. et al. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab. Eng.17 Oct 2009 (doi:10.1016/j.ymben.2009.10.003).
Vitkup, D., Kharchenko, P. & Wagner, A. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 7, R39 (2006).
Gianchandani, E. P., Papin, J. A., Price, N. D., Joyce, A. R. & Palsson, B. Ø. Matrix formalism to describe functional states of transcriptional regulatory systems. PLoS Comput. Biol. 2, e101 (2006).
Klamt, S., Saez-Rodriguez, J., Lindquist, J. A., Simeoni, L. & Gilles, E. D. A methodology for the structural and functional analysis of signaling and regulatory networks. BMC Bioinformatics 7, 56 (2006). This paper introduces the Boolean formalism for analysing large-scale signalling-network models.
Cho, B. K. et al. The transcription unit architecture of the Escherichia coli genome. Nature Biotech. 27, 1043–1049 (2009).
Hyduke, D. R., Jarboe, L. R., Tran, L. M., Chou, K. J. & Liao, J. C. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl Acad. Sci. USA 104, 8484–8489 (2007).
Jarboe, L. R., Hyduke, D. R., Tran, L. M., Chou, K. J. & Liao, J. C. Determination of the Escherichia coli S-nitrosoglutathione response network using integrated biochemical and systems analysis. J. Biol. Chem. 283, 5148–5157 (2008).
Shen-Orr, S. S., Milo, R., Mangan, S. & Alon, U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genet. 31, 64–68 (2002).
Balazsi, G., Barabasi, A. L. & Oltvai, Z. N. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc. Natl Acad. Sci. USA 102, 7841–7846 (2005).
Christensen, T. S., Oliveira, A. P. & Nielsen, J. Reconstruction and logical modeling of glucose repression signaling pathways in Saccharomyces cerevisiae. BMC Syst. Biol. 3, 7 (2009).
Papin, J. A. & Palsson, B. Ø. The JAK–STAT signaling network in the human B-cell: an extreme signaling pathway analysis. Biophys. J. 87, 37–46 (2004). An application of the stoichiometric approach that suggests that a substantial portion of the signalling pathways in the JAK–STAT pathway is not influenced by crosstalk.
Saez-Rodriguez, J. et al. A logical model provides insights into T cell receptor signaling. PLoS Comput. Biol. 3, e163 (2007). An application of the Boolean approach for analysing a real signalling pathway that includes experimental validation of a model prediction. It also shows how the Boolean approach can be used to identify gaps.
Sambrano, G. R. et al. Unravelling the signal-transduction network in B lymphocytes. Nature 420, 708–710 (2002).
Harmar, A. J. et al. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 37, D680–D685 (2009).
Olsen, J. V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006).
Peng, J. et al. A proteomics approach to understanding protein ubiquitination. Nature Biotech. 21, 921–926 (2003).
Zhou, W., Ryan, J. J. & Zhou, H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279, 32262–32268 (2004).
Jefferson, E. R., Walsh, T. P., Roberts, T. J. & Barton, G. J. SNAPPI-DB: a database and API of Structures, iNterfaces and Alignments for Protein–Protein Interactions. Nucleic Acids Res. 35, D580–D589 (2007).
Raghavachari, B., Tasneem, A., Przytycka, T. M. & Jothi, R. DOMINE: a database of protein domain interactions. Nucleic Acids Res. 36, D656–D661 (2008).
Breitkreutz, B. J. et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 36, D637–D640 (2008).
Papin, J. A. & Palsson, B. Ø. Topological analysis of mass-balanced signaling networks: a framework to obtain network properties including crosstalk. J. Theor. Biol. 227, 283–297 (2004).
Ernst, A. et al. Rapid evolution of functional complexity in a domain family. Sci. Signal. 2, ra50 (2009).
Gavin, A. C. et al. Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 (2006).
Manning, G., Whyte, D. B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Ubersax, J. A. & Ferrell, J. E. J. Mechanisms of specificity in protein phosphorylation. Nature Rev. Mol. Cell Biol. 8, 530–541 (2007).
Nijman, S. M. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 (2005).
Thiele, I., Jamshidi, N., Fleming, R. M. & Palsson, B. Ø. Genome-scale reconstruction of Escherichia coli's transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput. Biol. 5, e1000312 (2009).
Davidson, E. H. & Levine, M. S. Properties of developmental gene regulatory networks. Proc. Natl Acad. Sci. USA 105, 20063–20066 (2008).
Bhattacharyya, R. P., Remenyi, A., Yeh, B. J. & Lim, W. A. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75, 655–680 (2006).
Pawson, T. & Nash, P. Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 (2003).
Hsueh, R. C. et al. Deciphering signaling outcomes from a system of complex networks. Sci. Signal. 2, ra22 (2009). An experimental study of the response of macrophages to cytokines that shows how network properties can be deduced by treating the cell as a 'black box'.
Borisov, N. et al. Systems-level interactions between insulin–EGF networks amplify mitogenic signaling. Mol. Syst. Biol. 5, 256 (2009).
Sopko, R. & Andrews, B. J. Linking the kinome and phosphorylome — a comprehensive review of approaches to find kinase targets. Mol. Biosyst. 4, 920–933 (2008).
Skerker, J. M. et al. Rewiring the specificity of two-component signal transduction systems. Cell 133, 1043–1054 (2008).
Edwards, J. S. & Palsson, B. Ø. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc. Natl Acad. Sci. USA 97, 5528–5533 (2000).
Henry, C. S., Zinner, J. F., Cohoon, M. P. & Stevens, R. L. iBsu1103: a new genome-scale metabolic model of Bacillus subtilis based on SEED annotations. Genome Biol. 10, R69 (2009).
Kim, T. Y. et al. Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol. Bioeng. 97, 657–671 (2007).
Lee, D. S. et al. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J. Bacteriol. 191, 4015–4024 (2009).
Shinfuku, Y. et al. Development and experimental verification of a genome-scale metabolic model for Corynebacterium glutamicum. Microb. Cell Fact. 8, 43 (2009).
Suthers, P. F. et al. A genome-scale metabolic reconstruction of Mycoplasma genitalium, iPS189. PLoS Comput. Biol. 5, e1000285 (2009).
Thomas, G. H. et al. A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola. BMC Syst. Biol. 3, 24 (2009).
Chavali, A. K., Whittemore, J. D., Eddy, J. A., Williams, K. T. & Papin, J. A. Systems analysis of metabolism in the pathogenic trypanosomatid Leishmania major. Mol. Syst. Biol. 4, 177 (2008).
Sheikh, K., Forster, J. & Nielsen, L. K. Modeling hybridoma cell metabolism using a generic genome-scale metabolic model of Mus musculus. Biotechnol. Prog. 21, 112–121 (2005).
Notebaart, R. A., van Enckevort, F. H., Francke, C., Siezen, R. J. & Teusink, B. Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics 7, 296 (2006).
Oliveira, A. P., Patil, K. R. & Nielsen, J. Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst. Biol. 2, 17 (2008).
Zevedei-Oancea, I. & Schuster, S. A theoretical framework for detecting signal transfer routes in signalling networks. Comput. Chem. Eng. 29, 597–617 (2005).
Min Lee, J., Gianchandani, E. P., Eddy, J. A. & Papin, J. A. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput. Biol. 4, e1000086 (2008).
Kruger, M. et al. Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc. Natl Acad. Sci. USA 105, 2451–2456 (2008).
Luo, F. et al. Modular organization of protein interaction networks. Bioinformatics 23, 207–214 (2007).
Croes, D., Couche, F., Wodak, S. J. & van Helden, J. Metabolic PathFinding: inferring relevant pathways in biochemical networks. Nucleic Acids Res. 33, W326–W330 (2005).
Lin, C. Y. et al. Hubba: hub objects analyzer — a framework of interactome hubs identification for network biology. Nucleic Acids Res. 36, W438–W443 (2008).
Chang, R. L., Luo, F., Johnson, S. A. & Scheuermann, R. H. Deterministic graph-theoretic algorithm for detecting modules in biological interaction networks. Int. J. Bioinform. Res. Appl. (in the press).
Dasika, M. S., Burgard, A. & Maranas, C. D. A computational framework for the topological analysis and targeted disruption of signal transduction networks. Biophys. J. 91, 382–398 (2006).
Brady, A., Maxwell, K., Daniels, N. & Cowen, L. J. Fault tolerance in protein interaction networks: stable bipartite subgraphs and redundant pathways. PLoS ONE 4, e5364 (2009).
Jamshidi, N. & Palsson, B. Ø. Systems biology of SNPs. Mol. Syst. Biol. 2, 38 (2006).
Krantz, M. et al. Robustness and fragility in the yeast high osmolarity glycerol (HOG) signal-transduction pathway. Mol. Syst. Biol. 5, 281 (2009).
Mani, R., St Onge, R. P., Hartman, J. L., Giaever, G. & Roth, F. P. Defining genetic interaction. Proc. Natl Acad. Sci. USA 105, 3461–3466 (2008).
Sahin, O. et al. Combinatorial RNAi for quantitative protein network analysis. Proc. Natl Acad. Sci. USA 104, 6579–6584 (2007).
Green, M. L. & Karp, P. D. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics 5, 76 (2004).
Osterman, A. L. & Begley, T. P. A subsystems-based approach to the identification of drug targets in bacterial pathogens. Prog. Drug Res. 64, 131–170 (2007).
Kharchenko, P., Chen, L., Freund, Y., Vitkup, D. & Church, G. M. Identifying metabolic enzymes with multiple types of association evidence. BMC Bioinformatics 7, 177 (2006).
Reed, J. L. et al. Systems approach to refining genome annotation. Proc. Natl Acad. Sci. USA 103, 17480–17484 (2006).
Szczurek, E., Gat-Viks, I., Tiuryn, J. & Vingron, M. Elucidating regulatory mechanisms downstream of a signaling pathway using informative experiments. Mol. Syst. Biol. 5, 287 (2009). This paper introduces a creative experimental and theoretical approach that aims to expand signalling networks through targeted experiment design.
Shin, C. J., Wong, S., Davis, M. J. & Ragan, M. A. Protein–protein interaction as a predictor of subcellular location. BMC Syst. Biol. 3, 28 (2009).
Kelley, R. & Ideker, T. Systematic interpretation of genetic interactions using protein networks. Nature Biotech. 23, 561–566 (2005).
Bandyopadhyay, S., Kelley, R., Krogan, N. J. & Ideker, T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput. Biol. 4, e1000065 (2008).
Lehar, J. et al. Chemical combination effects predict connectivity in biological systems. Mol. Syst. Biol. 3, 80 (2007).
Venancio, T. M., Balaji, S., Iyer, L. M. & Aravind, L. Reconstructing the ubiquitin network: cross-talk with other systems and identification of novel functions. Genome Biol. 10, R33 (2009).
Singh, R., Xu, J. & Berger, B. Global alignment of multiple protein interaction networks with application to functional orthology detection. Proc. Natl Acad. Sci. USA 105, 12763–12768 (2008).
Kelley, B. P. et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc. Natl Acad. Sci. USA 100, 11394–11399 (2003).
Bandyopadhyay, S., Sharan, R. & Ideker, T. Systematic identification of functional orthologs based on protein network comparison. Genome Res. 16, 428–435 (2006).
Sharan, R. et al. Conserved patterns of protein interaction in multiple species. Proc. Natl Acad. Sci. USA 102, 1974–1979 (2005).
Tan, C. S. et al. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal. 2, ra39 (2009).
Cusick, M. E. et al. Literature-curated protein interaction datasets. Nature Methods 6, 39–46 (2009).
Crosson, S., McGrath, P. T., Stephens, C., McAdams, H. H. & Shapiro, L. Conserved modular design of an oxygen sensory/signaling network with species-specific output. Proc. Natl Acad. Sci. USA 102, 8018–8023 (2005).
Roguev, A. et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322, 405–410 (2008).
Beltrao, P. et al. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 7, e1000134 (2009).
Jacob, F. & Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 (1961).
Palsson, B. Systems Biology: Properties of Reconstructed Networks (Cambridge Univ. Press, 2006).
Covert, M. W., Knight, E. M., Reed, J. L., Herrgard, M. J. & Palsson, B. Ø. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429, 92–96 (2004).
Herrgard, M. J., Lee, B. S., Portnoy, V. & Palsson, B. Ø. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res. 16, 627–635 (2006).
Klipp, E., Nordlander, B., Kruger, R., Gennemark, P. & Hohmann, S. Integrative model of the response of yeast to osmotic shock. Nature Biotech. 23, 975–982 (2005).
Moxley, J. F. et al. Special feature: linking high-resolution metabolic flux phenotypes and transcriptional regulation in yeast modulated by the global regulator Gcn4p. Proc. Natl Acad. Sci. USA 106, 6477–6482 (2009).
Pryciak, P. M. Designing new cellular signaling pathways. Chem. Biol. 16, 249–254 (2009).
Antunes, M. S. et al. Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol. Syst. Biol. 5, 270 (2009).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Bulter, T. et al. Design of artificial cell–cell communication using gene and metabolic networks. Proc. Natl Acad. Sci. USA 101, 2299–2304 (2004).
Fung, E. et al. A synthetic gene-metabolic oscillator. Nature 435, 118–122 (2005).
Weber, W., Daoud-El Baba, M. & Fussenegger, M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc. Natl Acad. Sci. USA 104, 10435–10440 (2007).
Stricker, J. et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008).
Friedland, A. E. et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009).
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nature Biotech. 27, 753–759 (2009).
Acknowledgements
This work was supported in part by the US National Institute of Allergy and Infectious Diseases and the US Department of Health and Human Services through interagency agreement Y1-AI-8401-01.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Bernhard Ø. Palsson serves on the scientific advisory board of Genomatica, Inc.
Related links
Related links
DATABASES
The Alliance for Cellular Signaling
Database of Quantitative Cellular Signaling
DOMINE (database of protein domain interactions)
IUPHAR database of receptors and ion channels
Kyoto Encyclopedia of Genes and Genomes Pathway Database
SNAPPI (Structures, Interfaces and Alignments for Protein–Protein Interactions)
FURTHER INFORMATION
Glossary
- Biological network
-
A set of biological entities that act in an integrated fashion. Typical components of biological networks include organisms (for example, ecosystems, biofilms, and host–parasite or symbiont relationships), tissues (for example, the integrated operations of lungs, brain and heart), cells (for example, biofilms and different cells in a tissue) and the molecular components of cells (for example, proteins, DNA and RNA). Here, we focus on the molecular components of cells.
- Interactome
-
A set of molecular components of the cell, such as proteins, and the interactions between them. The interactions can be physical (protein A binds protein B) or correlative (perturbing protein A alters protein B's activity).
- Synthetic lethal
-
A genetic interaction in which the deletion of two genes at the same time results in lethality. An organism in which one gene is deleted and the other gene is present will still be viable.
- Formalism
-
A description of a property of a process in mathematical terms.
- Modules
-
A set of components that work in an integrated fashion. Personal computers are modular systems: they have keyboards, displays, motherboards and hard drives, each of which represents a module. Each module is relatively easy to replace but is composed of an integrated set of components that may be difficult to replace.
- Reverse engineering
-
The process of discovering the technological principles of a device, object or system through analysis of its structure, function and operation. It often involves taking a system apart and analysing its workings with the aim of making a new device or program that does the same thing without using any physical part of the original.
- Manual curation
-
In the context of this Review, this is the process by which a researcher assesses whether a given research paper possesses information that is relevant to a network under study and then examines the evidence put forth in the paper. This is opposed to artificial intelligence-guided text parsing or assuming a causal interaction based on correlative evidence in omics data sets.
- Serovar
-
A group of related microorganisms that are classified based on a characteristic set of antigens.
- Structural analysis
-
In the context of signalling networks, structural analysis focuses on exploring the connectivity of elements in the network and identifying routes from inputs to outputs. Structural analysis serves to construct a 'road map' of a network and then identify plausible routes from point A to point B. Connectivity refers to the number of different elements in the network with which a particular component may interact.
- Genetic interaction
-
An interaction in which one gene product alters the phenotypic effect of a second gene product. The most common phenotype of interest for genetic interaction studies is growth. Here, the interactions are observed through gene-deletion studies in which the growth phenotype of a single deletion is compared with the growth phenotype of a double deletion.
Rights and permissions
About this article
Cite this article
Hyduke, D., Palsson, B. Towards genome-scale signalling-network reconstructions. Nat Rev Genet 11, 297–307 (2010). https://doi.org/10.1038/nrg2750
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2750
This article is cited by
-
Translocating proteins compartment-specifically alter the fate of epithelial-mesenchymal transition in a compartmentalized Boolean network model
npj Systems Biology and Applications (2022)
-
Transcriptome analysis of the procession from chronic pancreatitis to pancreatic cancer and metastatic pancreatic cancer
Scientific Reports (2021)
-
The Key Genes of Chronic Pancreatitis which Bridge Chronic Pancreatitis and Pancreatic Cancer Can be Therapeutic Targets
Pathology & Oncology Research (2018)
-
Disentangling the multigenic and pleiotropic nature of molecular function
BMC Systems Biology (2015)
-
Large-scale models of signal propagation in human cells derived from discovery phosphoproteomic data
Nature Communications (2015)