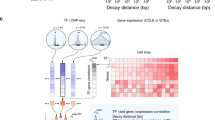
Key Points
-
Alterations in gene expression caused by the inappropriate level, structure or function of a transcription factor have been associated with a diverse set of human diseases. However, because most human transcription factors are essentially uncharacterized, the role of transcription factors in human health is currently greatly underappreciated.
-
Technological advances (such as chromatin immunoprecipitation followed by microarray or by sequencing) now allow transcription factor binding to be studied on a genome-wide scale.
-
Recent discoveries, such as the finding that most transcription factors bind to thousands of places in the genome, that binding sites are not just localized to proximal promoter regions and that some binding sites lack sequences similar to the consensus motif, have stimulated new ideas concerning long-range and combinatorial regulation.
-
Current genomic studies have not yet determined whether most human transcription factors bind alone or whether they cluster at hot spots in the genome. Answers to these questions require the genomic profiling of many more factors.
-
A crucial unanswered question is whether all binding events have a functional outcome (perhaps under some specific condition or in a specific cell type) or whether some transcription factor–genome interactions are simply irrelevant.
-
Issues that remain to be addressed include the design of comprehensive studies (for example, should all factors be studied in all cell types) and functional validation (for example, how can we determine the role of one specific binding site in its normal genomic context).
Abstract
A crucial question in the field of gene regulation is whether the location at which a transcription factor binds influences its effectiveness or the mechanism by which it regulates transcription. Comprehensive transcription factor binding maps are needed to address these issues, and genome-wide mapping is now possible thanks to the technological advances of ChIP–chip and ChIP–seq. This Review discusses how recent genomic profiling of transcription factors gives insight into how binding specificity is achieved and what features of chromatin influence the ability of transcription factors to interact with the genome. It also suggests future experiments that may further our understanding of the causes and consequences of transcription factor–genome interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Lee, T. I. & Young, R. A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34, 77–137 (2000). A detailed review of transcriptional regulation, general factors and accessory proteins that control transcription initation and elongation.
Sandelin, A. et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nature Rev. Genet. 8, 424–436 (2007).
ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007). This paper demonstrates how genome-wide studies of transcription factor binding, chromatin structure, DNA replication and sequence conservation can synergize.
Cooper, S. J., Trinklein, N. D., Anton, E. D., Nguyen, L. & Myers, R. M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 16, 1–10 (2006).
Kimura, K. et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 16, 55–65 (2006).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Vaquerizas, J. M., Kummerfeld, S. K., Teichmann, S. A. & Luscombe, N. M. A census of human transcription factors: function, expression and evolution. Nature Rev. Genet. 10, 252–263 (2009). A summary of the expression, conservation and activity of the set of human sequence-specific transcription factors.
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Jimenez-Sanchez, G., Childs, B. & Valle, D. Human disease genes. Nature 409, 853–855 (2001).
Wederell, E. D. et al. Global analysis of in vivo Foxa2-binding sites in mouse liver using massively parallel sequencing. Nucleic Acids Res. 36, 4549–4564 (2008).
Reed, B. D., Charos, A. E., Szekely, A. M., Weissman, S. M. & Snyder, M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLOS Genet. 4, e1000133 (2008).
Scacheri, P. C. et al. Genome-wide analysis of menin binding provides insights to MEN1 tumorigenesis. PLoS Genet. 2, e51 (2006).
Hatzis, P. et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 28, 2732–2744 (2008).
O'Geen, H. et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 3, e89 (2007).
Xu, X. et al. A comprehensive ChIP–chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals iterchangeable roles of E2F family members. Genome Res. 17, 1550–1561 (2007).
Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature Methods 4, 1–7 (2007). An early demonstration that high-throughput sequencing of ChIP samples can be used to identify genome-wide binding sites of site-specific transcription factors.
Johnson, D. S., Mortazavi, A., Myers, R. M. & Wold, B. Genome-wide mapping of in vivo protein–DNA interactions. Science 316, 1497–1502 (2007).
Rada-Iglesias, A. et al. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 18, 380–392 (2008).
Vogel, M. J., Peric-Hupkes, D. & van Steensel, B. Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nature Protoc. 2, 1467–1478 (2007).
Valouev, A. et al. Genome-wide analysis of transcription factor binding sites based on ChIP–seq data. Nature Methods 5, 829–834 (2008).
Johnson, D. S. et al. Systematic evaluation of variability in ChIP–chip experiments using predefined DNA targets. Genome Res. 18, 393–403 (2008).
Bieda, M., Xu, X., Singer, M., Green, R. & Farnham, P. J. Unbiased location analysis of E2F1 binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16, 595–605 (2006). A demonstration that some factors bind exclusively to proximal promoters and do not have strict motif requirements for their binding sites.
Kim, T. H. et al. A high-resolution map of active promoters in the human genome. Nature 436, 876–880 (2005). An early demonstration that high-density oligonucleotide arrays can be used to identify genome-wide binding sites for human transcription factors.
Cawley, S. et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116, 499–509 (2004).
Nix, D. A., Courdy, S. J. & Boucher, K. M. Empirical methods for controlling false positives and estimating confidence in ChIP–seq peaks. BMC Bioinformatics 9, 523 (2008).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Fejes, A. P. et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24, 1729–1730 (2008).
Rozowsky, J. et al. PeakSeq enables systematic scoring of ChIP–seq experiments relative to controls. Nature Biotech. 27, 66–75 (2009).
Liu, Y., Michalopoulos, G. K. & Zarnegar, R. Structural and functional characterization of the mouse hepatocyte growth factor gene promoter. J. Biol. Chem. 269, 4152–4160 (1994).
Fujishiro, K. et al. Analysis of tissue-specific and PPARα-dependent induction of FABP gene expression in the mouse liver by an in vivo DNA electroporation method. Mol. Cell. Biochem. 239, 165–172 (2002).
Weinmann, A. S., Yan, P. S., Oberley, M. J., Huang, T. H.-M. & Farnham, P. J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16, 235–244 (2002).
Li, Z. et al. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl Acad. Sci. USA 100, 8164–8169 (2003).
Ren, B. et al. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16, 245–256 (2002).
Odom, D. T. et al. Control of pancreas and liver gene expression by HNF transcription factors. Science 303, 1378–1381 (2004).
Consortium, T. E. P. The ENCODE (ENCyclopedia of DNA Elements) Project. Science 306, 636–640 (2004).
Carroll, J. S. et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005).
Yang, A. et al. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell 24, 593–602 (2006).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Mann, R. S. & Carroll, S. B. Molecular mechanisms of selector gene function and evolution. Curr. Opin. Genet. Dev. 12, 592–600 (2002).
Moorman, C. et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 12027–12032 (2006). This demonstrates clustering of transcription factors throughout the D. melanogaster genome.
Elnitski, L., Jin, V. X., Farnham, P. J. & Jones, S. J. M. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 16, 1455–1464 (2006).
Panne, D. The enhanceosome. Curr. Opin. Struct. Biol. 18, 236–242 (2008).
Maniatis, T. et al. Structure and function of the interferon-β enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 63, 609–620 (1998).
Dean, A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 22, 38–45 (2006).
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). This identifies specific histone modifications that are associated with cell-type-specific transcriptional regulation.
Heintzman, N. D. et al. Distinct predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genet. 39, 311–318 (2007).
Wright, W. E. & Funk, W. D. CASTing for multicomponent DNA-binding components. Trends Biochem. Sci. 18, 77–80 (1993).
Morgan, X. C., Ni, S., Miranker, D. P. & Iyer, V. R. Predicting combinatorial binding of transcription factors to regulatory elements in the human genome by association rule mining. BMC Bioinformatics 8, 445 (2007).
Rabinovich, A., Jin, V. X., Rabinovich, R., Xu, X. & Farnham, P. J. E2F in vivo binding specificity: comparison of consensus versus non-consensus binding sites. Genome Res. 18, 1763–1777 (2008).
Gineitis, D. & Treisman, R. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 276, 24531–24539 (2001).
Cooper, S. J., Trinklein, N. D., Nguyen, L. & Myers, R. M. Serum response factor binding sites differ in three human cell types. Genome Res. 17, 136–144 (2009).
Jin, V. X., O'Geen, H., Iyengar, S., Green, R. & Farnham, P. J. Identification of an OCT4 and SRY regulatory module using integrated computational and experimental genomics approaches. Genome Res. 17, 807–817 (2007).
Li, X.-Y. et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6, e27 (2008).
Badis, G. et al. Diversity and complexity in DNA recognition by transcription factors. Science 324, 1720–1723 (2009).
Li, H. et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 (2006).
Pena, P. V. et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 (2006).
Shi, X. et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282, 2450–2455 (2007).
Vermeulen, M. et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007).
Albert, T. et al. The chromatin structure of the dual c-myc promoter P1/P2 is regulated by separate elements. J. Biol. Chem. 276, 20482–20490 (2001).
Jin, V. X., Rabinovich, A., Squazzo, S. L., Green, R. & Farnham, P. J. A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data — a case study using E2F1. Genome Res. 16, 1585–1595 (2006).
Krig, S. R. et al. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)–chip assays. J. Biol. Chem. 282, 9703–9712 (2007).
Martone, R. et al. Distribution of NF-κB-binding sites across human chromosome 22. Proc. Natl Acad. Sci. USA 100, 12247–12252 (2003).
Krum, S. A. et al. Unique ERα cistromes control cell type-specific gene regulation. Mol. Endocrinol. 22, 2393–2406 (2008).
Voss, T. C. & Hager, G. L. Visualizing chromatin dynamics in intact cells. Biochem. Biophys. Acta 1783, 2044–2051 (2008).
Osborne, C. S. et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature Genet. 36, 1065–1071 (2004).
Bartlett, J. et al. Specialized transcription factories. Biochem. Soc. Symp. 73, 67–75 (2006).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Visel, A. et al. ChIP–seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009). This study demonstrates that using ChIP–seq to identify binding sites for p300 is a highly accurate method for identifying enhancers that can be shown, using follow-up assays in transgenic mice, to function in a tissue-specific manner.
Camenisch, T. D., Brilliant, M. H. & Segal, D. J. Critical parameters for genome editing using zinc finger nucleases. Mini Rev. Med. Chem. 8, 669–676 (2008).
Bletran, A., Liu, Y., Parikh, S., Temple, B. & Blancafort, P. Interrogating genomes with combinatorial artificial transcription factor libraries: asking zinc finger questions. Assay Drug Dev. Technol. 4, 317–331 (2006).
Faruqi, A. F., Egholm, M. & Glazer, P. M. Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cells. Proc. Natl Acad. Sci. USA 95, 1398–1403 (1998).
Burnett, R. et al. DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA-TCC repeats in Friedreich's ataxia. Proc. Natl Acad. Sci. USA 103, 11497–11502 (2006).
Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R. & Young, R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007).
Muse, G. W. et al. RNA polymerase is poised for activation across the genome. Nature Genet. 39, 1507–1511 (2007).
Komashko, V. M. et al. Using ChIP–chip technology to reveal common principles of transcriptional repression in normal and cancer cells. Genome Res. 18, 521–532 (2008).
Acevedo, L. G. et al. Genome-scale ChIP–chip analysis using 10,000 human cells. Biotechniques 43, 791–797 (2007).
O'Neill, L. P., VerMilyea, M. D. & Turner, B. M. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nature Genet. 38, 835–841 (2006).
Dahl, J. A. & Collas, P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells 25, 1037–1046 (2007).
Attema, J. L. et al. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc. Natl Acad. Sci. USA 104, 12371–12376 (2007).
Xu, H., Wei, C.-L., Lin, F. & Sung, W.-K. An HMM approach to genome-wide identification of differential histone modification sites from ChIP–seq data. Bioinformatics 24, 2344–2349 (2008).
Jothi, R., Cuddapah, S., Barski, A., Cui, K. & Zhao, K. Genome-wide identification of in vivo protein–DNA binding sites from ChIP–seq data. Nucleic Acids Res. 36, 5221–5231 (2008).
Hoffman, B. G. & Jones, S. J. Genome-wide identification of DNA–protein interactions using chromatin immunoprecipitation coupled with flow cell sequencing. J. Endocrinol. 201, 1–13 (2009).
Acknowledgements
The author thanks X. Xu, H. O'Geen and S. Frietze for providing data used in figure 2 and the members of the Farnham laboratory for their insights and discussions.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
Peggy J. Farnham is a member of the ENCODE Consortium and a member of an Epigenome Mapping Center, both of which are mentioned in the Review.
Related links
Glossary
- TATA box
-
A consensus sequence in promoters that is enriched in thymine and adenine residues and is important for the recruitment of the general transcriptional machinery at some promoters.
- Initiator
-
An element with a consensus of YYANWYY (in which A is the transcription start site, N is any nucleotide, W is adenosine or thymine, and Y is a pyrimidine) that helps to recruit the general transcriptional machinery to promoters.
- Initiation complex
-
The assembly of RNA polymerase and associated general factors that binds to the core promoter region.
- Silencer
-
A DNA sequence capable of binding transcription factors that are termed repressors, which can negatively influence transcription by preventing recruitment of the general transcriptional machinery or by recruiting histone-modifying complexes that create repressive chromatin structures.
- Transcription factor II D
-
A protein complex composed of several subunits, called TATA binding protein (TBP)-associated factors (TAFs), and the TBP. It is one of several complexes that make up the RNA polymerase II initiation machinery.
- DamID
-
An alternative method to chromatin immunoprecipitation that uses a DNA-binding protein fused to a DNA methyltransferase. Adenine methylation of a region identifies it as being located near a binding site.
- Reporter construct
-
A plasmid containing a promoter (and sometimes an enhancer) cloned upstream of a reporter gene (often simply called the reporter) that is introduced into cultured cells, animals or plants. Certain genes are chosen as reporters because their products can be easily or quantitatively assayed, or used as selectable markers.
- CpG island
-
A sequence of at least 200 bp with a greater number of CpG sites than expected for its GC content. These regions are often GC rich and usually undermethylated. They correspond to the promoter regions of many mammalian genes.
- Enhanceosome
-
A protein complex that binds to an enhancer region (which can be located upstream, downstream or in a gene); the transcription factors that compose the enhanceosome are thought to work cooperatively to stimulate transcription.
- Heterochromatin
-
Chromatin that is characterized by very dense packing of DNA, which makes it less accessible to transcription factors. Certain regions of the genome, such as centromeres and telomeres, are always heterochromatinized (constitutive heterochromatin regions), whereas other regions are densely packed and repressed only in certain cells (facultative heterochromatin regions).
- DNA methylation
-
An epigenetic DNA modification that can be added and removed without changing the original DNA sequence and that is characterized by the addition of a methyl group to the number 5 carbon of the cytosine pyrimidine ring.
- Plant homeodomain finger
-
A 50–80 amino acid domain that contains a Cys4-His-Cys3 motif. It is found in more than 100 human proteins, several of which are involved in chromatin-mediated gene regulation.
- Small interfering RNAs
-
Small antisense RNAs (20–25 nucleotides) that can be directly introduced into cells or be generated in cells from longer dsRNAs. They serve as guides for the cleavage of homologous mRNA in the RNA-induced silencing complex.
- Transcription factory
-
A nuclear subcompartment that is rich in RNA polymerases and transcription factors, and in which there is clustering of active genes.
- Interactome
-
A complete set of macromolecular interactions (physical and genetic). Current use of the word tends to refer to a comprehensive set of protein–protein interactions. However, the protein–DNA interactome (a network formed by transcription factors and their target genes) is also commonly studied.
- Artificial zinc finger
-
Chimaeras of zinc finger domains — small protein domains that coordinate one or more zinc ions and that are commonly found in mammalian transcription factors — and an effector domain (for example, an activator, repressor, methylase or nuclease). Linking together six zinc fingers produces a target site of 18 bp, which is long enough to be unique in all known genomes.
Rights and permissions
About this article
Cite this article
Farnham, P. Insights from genomic profiling of transcription factors. Nat Rev Genet 10, 605–616 (2009). https://doi.org/10.1038/nrg2636
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2636
This article is cited by
-
An Overview of Molecular Basis and Genetic Modification of Floral Organs Genes: Impact of Next-Generation Sequencing
Molecular Biotechnology (2023)
-
CeTF: an R/Bioconductor package for transcription factor co-expression networks using regulatory impact factors (RIF) and partial correlation and information (PCIT) analysis
BMC Genomics (2021)
-
Molecular and computational approaches to map regulatory elements in 3D chromatin structure
Epigenetics & Chromatin (2021)
-
Constructing gene regulatory networks using epigenetic data
npj Systems Biology and Applications (2021)
-
Gene co-regulation and co-expression in the aryl hydrocarbon receptor-mediated transcriptional regulatory network in the mouse liver
Archives of Toxicology (2020)