Key Points
-
Epigenetic processes have been implicated in the pathogenesis of type 2 diabetes mellitus
-
Diet and exercise might affect the epigenome over several generations
-
Epigenetic changes can be driven by DNA methylation and histone modification in response to environmental stressors
-
Regulation of gene expression by DNA methylation and histone modification occurs by a mechanism that impairs the access of transcriptional machinery to the promoters
-
Studying the epigenetic signatures of insulin resistance and the adaptive response to exercise might provide insight into gene–environment networks that control glucose and energy homeostasis.
Abstract
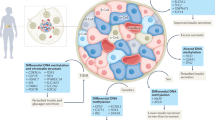
Epigenetic changes are caused by biochemical regulators of gene expression that can be transferred across generations or through cell division. Epigenetic modifications can arise from a variety of environmental exposures including undernutrition, obesity, physical activity, stress and toxins. Transient epigenetic changes across the entire genome can influence metabolic outcomes and might or might not be heritable. These modifications direct and maintain the cell-type specific gene expression state. Transient epigenetic changes can be driven by DNA methylation and histone modification in response to environmental stressors. A detailed understanding of the epigenetic signatures of insulin resistance and the adaptive response to exercise might identify new therapeutic targets that can be further developed to improve insulin sensitivity and prevent obesity. This Review focuses on the current understanding of mechanisms by which lifestyle factors affect the epigenetic landscape in type 2 diabetes mellitus and obesity. Evidence from the past few years about the potential mechanisms by which diet and exercise affect the epigenome over several generations is discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
World Health Organization. Global report on diabetes. http://www.who.int/diabetes/global-report/en/ (2006).
Wilcox, G. Insulin and insulin resistance. Clin. Biochem. Rev. 26, 19–39 (2005).
Moller, D. E. & Kaufman, K. D. Metabolic syndrome: a clinical and molecular perspective. Annu. Rev. Med. 56, 45–62 (2005).
Franks, P. W., Pearson, E. & Florez, J. C. Gene–environment and gene–treatment interactions in type 2 diabetes: progress, pitfalls, and prospects. Diabetes Care 36, 1413–1421 (2013).
Billings, L. K. & Florez, J. C. The genetics of type 2 diabetes: what have we learned from GWAS? Ann. NY Acad. Sci. 1212, 59–77 (2010).
Bonnefond, A. & Froguel, P. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metab. 21, 357–368 (2015).
Prasad, R. B. & Groop, L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 6, 87–123 (2015).
McCarthy, M. I. Genomic medicine at the heart of diabetes management. Diabetologia 58, 1725–1729 (2015).
Groop, L. & Pociot, F. Genetics of diabetes — are we missing the genes or the disease? Mol. Cell. Endocrinol. 382, 726–739 (2014).
Kirchner, H., Osler, M. E., Krook, A. & Zierath, J. R. Epigenetic flexibility in metabolic regulation: disease cause and prevention? Trends Cell Biol. 23, 203–209 (2013).
Ling, C. & Groop, L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 58, 2718–2725 (2009).
Ong, T. P. & Ozanne, S. E. Developmental programming of type 2 diabetes: early nutrition and epigenetic mechanisms. Curr. Opin. Clin. Nutr. Metab. Care 18, 354–360 (2015).
Tracey, R., Manikkam, M., Guerrero-Bosagna, C. & Skinner, M. K. Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. Reprod. Toxicol. 36, 104–116 (2013).
Bird, A. Perceptions of epigenetics. Nature 447, 396–398 (2007).
Costa, F. F. Epigenomics in cancer management. Cancer Manag. Res. 2, 255–265 (2010).
Youngson, N. A. & Whitelaw, E. Transgenerational epigenetic effects. Annu. Rev. Genomics Hum. Genet. 9, 233–257 (2008).
Pembrey, M. E. et al. Sex-specific, male-line transgenerational responses in humans. Eur. J. Hum. Genet. 14, 159–166 (2006).
Fernandez-Twinn, D. S. et al. Maternal protein restriction leads to hyperinsulinemia and reduced insulin-signaling protein expression in 21-mo-old female rat offspring. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R368–R373 (2005).
Hales, C. N. & Barker, D. J. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia 35, 595–601 (1992).
Hales, C. N. & Barker, D. J. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. 1992. Int. J. Epidemiol. 42, 1215–1222 (2013).
Ozanne, S. E., Sandovici, I. & Constancia, M. Maternal diet, aging and diabetes meet at a chromatin loop. Aging (Albany NY) 3, 548–554 (2011).
Petry, C. J., Dorling, M. W., Pawlak, D. B., Ozanne, S. E. & Hales, C. N. Diabetes in old male offspring of rat dams fed a reduced protein diet. Int. J. Exp. Diabetes Res. 2, 139–143 (2001).
Sandovici, I. et al. Maternal diet and aging alter the epigenetic control of a promoter–enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc. Natl Acad. Sci. USA 108, 5449–5454 (2011).
Dahri, S., Snoeck, A., Reusens-Billen, B., Remacle, C. & Hoet, J. J. Islet function in offspring of mothers on low-protein diet during gestation. Diabetes 40 (Suppl. 2), 115–120 (1991).
Ravelli, A. C. et al. Glucose tolerance in adults after prenatal exposure to famine. Lancet 351, 173–177 (1998).
Lumey, L. H. et al. Cohort profile: the Dutch Hunger Winter families study. Int. J. Epidemiol. 36, 1196–1204 (2007).
Li, Y. et al. Exposure to the Chinese famine in early life and the risk of hyperglycemia and type 2 diabetes in adulthood. Diabetes 59, 2400–2406 (2010).
Lumey, L. H., Khalangot, M. D. & Vaiserman, A. M. Association between type 2 diabetes and prenatal exposure to the Ukraine famine of 1932–33: a retrospective cohort study. Lancet Diabetes Endocrinol. 3, 787–794 (2015).
Thurner, S. et al. Quantification of excess risk for diabetes for those born in times of hunger, in an entire population of a nation, across a century. Proc. Natl Acad. Sci. USA 110, 4703–4707 (2013).
Kaati, G., Bygren, L. O. & Edvinsson, S. Cardiovascular and diabetes mortality determined by nutrition during parents' and grandparents' slow growth period. Eur. J. Hum. Genet. 10, 682–688 (2002).
Faulk, C., Barks, A., Liu, K., Goodrich, J. M. & Dolinoy, D. C. Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 5, 487–500 (2013).
de Castro Barbosa, T. et al. High-fat diet reprograms the epigenome of rat spermatozoa and transgenerationally affects metabolism of the offspring. Mol. Metab. 5, 184–197 (2016).
Ng, S. F. et al. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467, 963–966 (2010).
Warram, J. H., Krolewski, A. S., Gottlieb, M. S. & Kahn, C. R. Differences in risk of insulin-dependent diabetes in offspring of diabetic mothers and diabetic fathers. N. Engl. J. Med. 311, 149–152 (1984).
Lee, J. T. & Bartolomei, M. S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308–1323 (2013).
Weaver, I. C. et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 (2004).
Drake, A. J. et al. Reduced adipose glucocorticoid reactivation and increased hepatic glucocorticoid clearance as an early adaptation to high-fat feeding in Wistar rats. Endocrinology 146, 913–919 (2005).
Hardikar, A. A. et al. Multigenerational undernutrition increases susceptibility to obesity and diabetes that is not reversed after dietary recuperation. Cell Metab. 22, 312–319 (2015).
Dunn, G. A. & Bale, T. L. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology 150, 4999–5009 (2009).
Yokomizo, H. et al. Maternal high-fat diet induces insulin resistance and deterioration of pancreatic β-cell function in adult offspring with sex differences in mice. Am. J. Physiol. Endocrinol. Metab. 306, E1163–E1175 (2014).
Wei, Y. et al. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc. Natl Acad. Sci. USA 111, 1873–1878 (2014).
Fullston, T. et al. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 27, 4226–4243 (2013).
Buescher, J. L. et al. Evidence for transgenerational metabolic programming in Drosophila. Dis. Model. Mech. 6, 1123–1132 (2013).
Carone, B. R. et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143, 1084–1096 (2010).
Radford, E. J. et al. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 345, 1255903 (2014).
Skinner, M. K. et al. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med. 11, 228 (2013).
Sanford, J. P., Clark, H. J., Chapman, V. M. & Rossant, J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1, 1039–1046 (1987).
Aiken, C. E., Tarry-Adkins, J. L. & Ozanne, S. E. Transgenerational developmental programming of ovarian reserve. Sci. Rep. 5, 16175 (2015).
Terashima, M. et al. Effect of high fat diet on paternal sperm histone distribution and male offspring liver gene expression. Epigenetics 10, 861–871 (2015).
Egan, B. & Zierath, J. R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17, 162–184 (2013).
Hawley, J. A., Hargreaves, M., Joyner, M. J. & Zierath, J. R. Integrative biology of exercise. Cell 159, 738–749 (2014).
[No authors listed.] Impact of physical activity during pregnancy and postpartum on chronic disease risk. Med. Sci. Sports Exerc. 38, 989–1006 (2006).
Mourtakos, S. P. et al. Maternal lifestyle characteristics during pregnancy, and the risk of obesity in the offspring: a study of 5,125 children. BMC Pregnancy Childbirth 15, 66 (2015).
Laker, R. C. et al. Exercise prevents maternal high-fat diet-induced hypermethylation of the Pgc-1α gene and age-dependent metabolic dysfunction in the offspring. Diabetes 63, 1605–1611 (2014).
Lin, J., Handschin, C. & Spiegelman, B. M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1, 361–370 (2005).
Stanford, K. I. et al. Exercise before and during pregnancy prevents the deleterious effects of maternal high-fat feeding on metabolic health of male offspring. Diabetes 64, 427–433 (2015).
Sheldon, R. D. et al. Gestational exercise protects adult male offspring from high-fat diet-induced hepatic steatosis. J. Hepatol. 64, 171–178 (2016).
Murashov, A. K. et al. Paternal long-term exercise programs offspring for low energy expenditure and increased risk for obesity in mice. FASEB J. 30, 775–784 (2016).
Carter, L. G., Qi, N. R., De Cabo, R. & Pearson, K. J. Maternal exercise improves insulin sensitivity in mature rat offspring. Med. Sci. Sports Exerc. 45, 832–840 (2013).
Guth, L. M. et al. Sex-specific effects of exercise ancestry on metabolic, morphological and gene expression phenotypes in multiple generations of mouse offspring. Exp. Physiol. 98, 1469–1484 (2013).
McPherson, N. O., Owens, J. A., Fullston, T. & Lane, M. Preconception diet or exercise intervention in obese fathers normalizes sperm microRNA profile and metabolic syndrome in female offspring. Am. J. Physiol. Endocrinol. Metab. 308, E805–E821 (2015).
Zierath, J. R. & Wallberg-Henriksson, H. Looking ahead perspective: where will the future of exercise biology take us? Cell Metab. 22, 25–30 (2015).
Donkin, I. et al. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab. 23, 369–378 (2016).
Denham, J., O'Brien, B. J., Harvey, J. T. & Charchar, F. J. Genome-wide sperm DNA methylation changes after 3 months of exercise training in humans. Epigenomics 7, 1–15 (2015).
Dalgaard, K. et al. Trim28 haploinsufficiency triggers bi-stable epigenetic obesity. Cell 164, 353–364 (2016).
Vaag, A. & Poulsen, P. Twins in metabolic and diabetes research: what do they tell us? Curr. Opin. Clin. Nutr. Metab. Care 10, 591–596 (2007).
Fraga, M. F. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA 102, 10604–10609 (2005).
Martin, G. M. Epigenetic drift in aging identical twins. Proc. Natl Acad. Sci. USA 102, 10413–10414 (2005).
Ollikainen, M. et al. Genome-wide blood DNA methylation alterations at regulatory elements and heterochromatic regions in monozygotic twins discordant for obesity and liver fat. Clin. Epigenetics 7, 39 (2015).
Zhao, J., Goldberg, J., Bremner, J. D. & Vaccarino, V. Global DNA methylation is associated with insulin resistance: a monozygotic twin study. Diabetes 61, 542–546 (2012).
Yuan, W. et al. An integrated epigenomic analysis for type 2 diabetes susceptibility loci in monozygotic twins. Nat. Commun. 5, 5719 (2014).
Ribel-Madsen, R. et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS ONE 7, e51302 (2012).
Nilsson, E. et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 63, 2962–2976 (2014).
Pietilainen, K. H. et al. DNA methylation and gene expression patterns in adipose tissue differ significantly within young adult monozygotic BMI-discordant twin pairs. Int. J. Obes. (Lond.) 40, 654–661 (2016).
Dayeh, T. et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 10, e1004160 (2014).
Volkmar, M. et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 31, 1405–1426 (2012).
Yang, B. T. et al. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol. Endocrinol. 26, 1203–1212 (2012).
Stitzel, M. L. et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 12, 443–455 (2010).
Barres, R. et al. Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 10, 189–198 (2009).
Kulkarni, S. S. et al. Mitochondrial regulators of fatty acid metabolism reflect metabolic dysfunction in type 2 diabetes mellitus. Metabolism 61, 175–185 (2012).
Barres, R. et al. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 3, 1020–1027 (2013).
Multhaup, M. L. et al. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metab. 21, 138–149 (2015).
Nilsson, E. et al. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J. Clin. Endocrinol. Metab. 100, E1491–E1501 (2015).
Kirchner, H. et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol. Metab. 5, 171–183 (2016).
Xu, X. et al. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics 8, 522–533 (2013).
Agha, G. et al. Adiposity is associated with DNA methylation profile in adipose tissue. Int. J. Epidemiol. 44, 1277–1287 (2015).
Guenard, F. et al. Differential methylation in visceral adipose tissue of obese men discordant for metabolic disturbances. Physiol. Genomics 46, 216–222 (2014).
Keller, M. et al. Global DNA methylation levels in human adipose tissue are related to fat distribution and glucose homeostasis. Diabetologia 57, 2374–2383 (2014).
Horvath, S. et al. Obesity accelerates epigenetic aging of human liver. Proc. Natl Acad. Sci. USA 111, 15538–15543 (2014).
El-Osta, A. et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 205, 2409–2417 (2008).
Hall, E. et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC Med. 12, 103 (2014).
Ishikawa, K. et al. Long-term pancreatic beta cell exposure to high levels of glucose but not palmitate induces DNA methylation within the insulin gene promoter and represses transcriptional activity. PLoS ONE 10, e0115350 (2015).
Pirola, L. et al. Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res. 21, 1601–1615 (2011).
Jacobsen, S. C. et al. Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia 55, 3341–3349 (2012).
Dahlman, I. et al. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. Int. J. Obes. (Lond.) 39, 910–919 (2015).
Barres, R. et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 15, 405–411 (2012).
Yu, M. et al. Metabolic and mitogenic signal transduction in human skeletal muscle after intense cycling exercise. J. Physiol. 546, 327–335 (2003).
McGee, S. L. & Hargreaves, M. Exercise and myocyte enhancer factor 2 regulation in human skeletal muscle. Diabetes 53, 1208–1214 (2004).
McGee, S. L., Fairlie, E., Garnham, A. P. & Hargreaves, M. Exercise-induced histone modifications in human skeletal muscle. J. Physiol. 587, 5951–5958 (2009).
Nitert, M. D. et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 61, 3322–3332 (2012).
Rowlands, D. S. et al. Multi-omic integrated networks connect DNA methylation and miRNA with skeletal muscle plasticity to chronic exercise in type 2 diabetic obesity. Physiol. Genomics 46, 747–765 (2014).
Mukwevho, E. et al. Caffeine induces hyperacetylation of histones at the MEF2 site on the Glut4 promoter and increases MEF2A binding to the site via a CaMK-dependent mechanism. Am. J. Physiol. Endocrinol. Metab. 294, E582–E588 (2008).
Ronn, T. et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 9, e1003572 (2013).
Aoi, W. et al. The microRNA miR-696 regulates PGC-1α in mouse skeletal muscle in response to physical activity. Am. J. Physiol. Endocrinol. Metab. 298, E799–E806 (2010).
Nielsen, S. et al. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 588, 4029–4037 (2010).
Russell, A. P. et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 591, 4637–4653 (2013).
Tonevitsky, A. G. et al. Dynamically regulated miRNA–mRNA networks revealed by exercise. BMC Physiol. 13, 9 (2013).
de Gonzalo-Calvo, D. et al. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J. Appl. Physiol. (1985) 119, 124–134 (2015).
Parrizas, M. et al. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J. Clin. Endocrinol. Metab. 100, E407–E415 (2015).
Wardle, S. L. et al. Plasma microRNA levels differ between endurance and strength athletes. PLoS ONE 10, e0122107 (2015).
McKinsey, T. A., Zhang, C. L., Lu, J. & Olson, E. N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106–111 (2000).
Smith, J. A., Kohn, T. A., Chetty, A. K. & Ojuka, E. O. CaMK activation during exercise is required for histone hyperacetylation and MEF2A binding at the MEF2 site on the Glut4 gene. Am. J. Physiol. Endocrinol. Metab. 295, E698–E704 (2008).
Acknowledgements
The authors are supported by grants to R.B. and J.R.Z. from the Novo Nordisk Foundation, and to J.R.Z. from the Swedish Research Council, The European Research Council and the Strategic Program in Diabetes Research at Karolinska Institutet.
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article, contributed to discussion of the content, wrote the article and reviewed and/or edited the article before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Barrès, R., Zierath, J. The role of diet and exercise in the transgenerational epigenetic landscape of T2DM. Nat Rev Endocrinol 12, 441–451 (2016). https://doi.org/10.1038/nrendo.2016.87
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrendo.2016.87
This article is cited by
-
The influence of insulin-related genetic variants on fetal growth, fetal blood flow, and placental weight in a prospective pregnancy cohort
Scientific Reports (2023)
-
DNA hypomethylation characterizes genes encoding tissue-dominant functional proteins in liver and skeletal muscle
Scientific Reports (2023)
-
The association between dietary patterns derived by three statistical methods and type 2 diabetes risk: YaHS-TAMYZ and Shahedieh cohort studies
Scientific Reports (2023)
-
Epigenetic modifications influence urinary tract infection outcome
Nature Microbiology (2023)
-
FHL3 promotes the formation of fast glycolytic muscle fibers by interacting with YY1 and muscle glycolytic metabolism
Cellular and Molecular Life Sciences (2023)