Abstract
Phenotypic drug discovery (PDD) approaches do not rely on knowledge of the identity of a specific drug target or a hypothesis about its role in disease, in contrast to the target-based strategies that have been widely used in the pharmaceutical industry in the past three decades. However, in recent years, there has been a resurgence in interest in PDD approaches based on their potential to address the incompletely understood complexity of diseases and their promise of delivering first-in-class drugs, as well as major advances in the tools for cell-based phenotypic screening. Nevertheless, PDD approaches also have considerable challenges, such as hit validation and target deconvolution. This article focuses on the lessons learned by researchers engaged in PDD in the pharmaceutical industry and considers the impact of 'omics' knowledge in defining a cellular disease phenotype in the era of precision medicine, introducing the concept of a chain of translatability. We particularly aim to identify features and areas in which PDD can best deliver value to drug discovery portfolios and can contribute to the identification and the development of novel medicines, and to illustrate the challenges and uncertainties that are associated with PDD in order to help set realistic expectations with regard to its benefits and costs.
Similar content being viewed by others
Main
In the past three decades, target-based drug discovery (TDD) — in which the starting point is a defined molecular target that is hypothesized to have an important role in disease — has been the dominant approach to drug discovery in the pharmaceutical industry, driven by advances in molecular biology and genomics. However, in recent years, there has been a revival in interest in phenotypic drug discovery (PDD) approaches, which do not rely on knowledge of the identity of a specific drug target or a hypothesis about its role in disease.
This interest has been catalysed in part by an influential analysis by Swinney and Anthony in 2011 that highlighted the strong contribution of PDD to the discovery of first-in-class drugs1. PDD has since been cited by various authors as a potential solution to the perceived poor productivity of TDD (for example, see Ref. 2), and from the authors' experience and anecdotal communications, it seems that efforts within the pharmaceutical industry to pursue PDD have recently greatly increased compared with the years preceding 2011. The power of PDD as a tool to address the complexity of diseases that are poorly understood by the scientific community is also clear (see Ref. 3 for a recent review). Furthermore, there have been recent rapid advances in various technologies for cell-based phenotypic screening, including the development of induced pluripotent stem (iPS) cell technologies4, gene-editing tools such as CRISPR–Cas5, organoids and imaging assay technologies. Such tools have enabled the development of novel cell-based disease models that promise to more realistically recapitulate human disease biology.
Nevertheless, there are still challenges in prospectively understanding the key success factors for modern PDD and how maximal value can be obtained. Articles published after the analysis by Swinney and Anthony have re-examined the contribution of PDD to new drug discovery6,7 and have refined the conditions for its successful application8. Importantly, it is apparent on closer examination that the classification of drugs as 'phenotypically discovered' is somewhat inconsistent6,7 and that, in fact, the majority of successful drug discovery programmes combine target knowledge and functional cellular assays to identify drug candidates with the most advantageous molecular mechanism of action (MoA). Although there is clear evidence that phenotypic screening can be an attractive proposition for efficiently identifying functionally active hits that lead to first-in-class drugs, the gap between a screening hit and an efficacious drug is often immense and, in our experience, more challenging than for a hit with a known molecular target. Hopes for PDD to 'rescue' the pharmaceutical industry might also be viewed as an example of a Gartner hype cycle, in which a peak of inflated expectations is followed by a trough of disillusionment, before establishing a plateau of productivity.
This article aims to address two aspects of this situation: first, to illustrate current challenges and uncertainties that are associated with PDD to set realistic expectations for benefits and costs; and second, to identify areas in which PDD can best deliver value to drug discovery portfolios through the identification and the development of novel medicines. In the past two years, conferences organized by the New York Academy of Sciences9 and the Keystone Symposia on Molecular and Cellular Biology, along with the establishment of a PDD special interest group under the auspices of the Society for Laboratory Automation and Screening, have sustained discussion around PDD and its value for the pharmaceutical industry. This article also aims to capture the spirit of these recent meetings by focusing on the state of the art in PDD, sharing lessons learned, and carefully examining the opportunities and challenges for PDD. We first highlight core concepts in PDD and introduce the concept of a chain of translatability for PDD screens, and then discuss strategic considerations and operational aspects for PDD projects, including library development, hit triage, compound optimization and safety assessments.
Core concepts in PDD
Defining PDD. Drugs typically act by engaging a molecular target; however, a priori knowledge of that target is not essential. In the case of PDD, a 'physiologically relevant' biological system or cellular signalling pathway is directly interrogated by chemical matter to identify biologically active compounds. This target-agnostic approach is the underlying attribute that differentiates PDD from hypothesis-driven TDD10. These target-agnostic and empirical aspects of PDD are consistent with its description and usage by scientists in academia and industry. Use of a uniform definition for PDD helps to illuminate the impact of PDD on modern drug discovery1,6, and underlines the importance and impact of empirical drug discovery approaches in an era that is dominated by strategies that are based on molecular target hypotheses1. Although primarily an approach for small-molecule discovery, PDD has also contributed to antibody drug discovery (see the excellent review by Gonzalez-Munoz et al.11).
Most drug discovery projects that are based on a molecular target hypothesis also test active compounds in phenotypic cellular assays. Although these are clearly not phenotypic or empirical drug discovery examples, novel and therapeutically important MoAs that differentiate targeted drugs can be discovered phenotypically. An example of this 'molecularly informed phenotypic discovery' paradigm3 was the empirical observation that the oestrogen receptor (ER) antagonist fulvestrant displayed greater than expected efficacy, leading to the elucidation of its ER-degrading mechanism.
Eder et al.6 noted that many drugs discovered by target-unbiased empirical pharmacology, which could potentially be called 'phenotypically discovered' drugs, were the outcome of serendipitous observations or of the individual phenotypic characterization of known active pharmacophores, rather than a result of a disease-first lead discovery effort. Going forwards, in order to substantially improve industry productivity, we argue that PDD must proceed rationally from disease understanding to a mechanistically defined effect on a pathway or a biomarker to drug, and then to a therapeutic effect.
Chain of translatability. The fundamental determinant of the potential success of a PDD effort is the ability of the screening assay to predict the clinical therapeutic response to a drug with a specific mechanism of action. This was described by Scannell and Bosley as the “predictive validity” of a discovery model2. Here, we propose the term chain of translatability to describe the presence of a shared mechanistic basis for the disease model, the assay readout and the biology of the disease in humans, as a framework for developing phenotypic screening assays with a greater likelihood of having strong predictive validity.
PDD projects in the area of infectious disease (seeking antibiotics12, antivirals13 and anti-parasitic agents14) often have a strong chain of translatability. Indeed, a typical PDD assay readout — inhibiting the replication of bacteria, viruses or parasites on plates — can strongly correspond not only to anti-infective activity in in vivo preclinical models, but also to the pharmacodynamic (PD) and the therapeutic effects sought in patients. For example, the anti-hepatitis C virus (HCV) drug daclatasvir was discovered phenotypically using human cells engineered to express the HCV replicon corresponding to a number of clinically relevant genotypes15. Therefore, such a system was strongly predictive for the inhibition of HCV replication in vivo. The molecular target of daclatasvir and of subsequent anti-HCV drugs, the HCV NS5A protein, was not previously regarded as a target as it then had no known function.
Similarly, PDD projects that aim to modulate the production of proteins with either known human pharmacological activity (for example, insulin) or a highly validated association with human physiology (for example, PCSK9 (Ref. 16)) can have a strong chain of translatability. Based on human genetics, there was a very strong rationale that the reduction of PCSK9 synthesis would have beneficial effects in lowering LDL cholesterol levels. A screen for agents that reduced PCSK9 production in CHO cells17 identified a totally novel molecular MoA — gene-specific ribosome stalling. As the compounds act upon such a fundamental and conserved mechanism, there is a very high probability that, given a molecule with appropriate pharmaceutical properties, the same molecular MoA would also be active in vivo.
Rare monogenic diseases can also provide an opportunity to establish PDD projects with a strong chain of translatability18,19, as knowledge of the mutation that causes the disease can be used as the basis for the development of disease models that are suitable for PDD. For example, pioneering genome-wide expression analysis in patients with recessive dystrophic epidermolysis bullosa (RDEB), a rare disease that is characterized by fragile skin due to mutations in the COL7A1 gene, showed that the differential expression of genes associated with the transforming growth factor-β (TGFβ) pathway was responsible for differences in the clinical manifestation of the disease. Based on these results, and the knowledge that the approved angiotensin II receptor antagonist losartan attenuates both canonical and non-canonical TGFβ signalling, Nyström et al.20 demonstrated that long-term losartan treatment of COL7A1-mutant RDEB mice efficiently reduced TGFβ signalling in chronically injured forepaws and alleviated hallmarks of RDEB progression. It is possible that a differential phenotypic screen using wild-type and COL7A1-deficient fibroblasts could identify compounds that modulate or that prevent RDEB disease progression to a greater extent than losartan. As in the case of antibacterial and anti-parasitic drugs, the chain of translatability for monogenic diseases may extend not only from effects in a cell-based phenotypic assay to effects in a preclinical animal model, but also to therapeutic effects in humans, as the same mutation found in humans frequently often drives a very comparable phenotype, disease natural history and outcome in animal models that have, or that are anticipated to have, good predictive validity.
Beyond the strong potential for PDD to contribute substantially to the identification of therapeutics for patients with some of the 7,000 known rare diseases (many of which have no specific treatments), such efforts could also have implications for broader patient populations. Rare diseases are in many cases highly suitable for the evaluation, within a very homogenous (although small) patient population, of a novel MoA that might be relevant in other, more prevalent indications that share key disease characteristics21.
Unfortunately, there are many therapeutic indications that have neither a highly predictive animal model nor a quantifiable phenotypic assay end point that mechanistically corresponds to a causal disease biomarker. Oncology and neuroscience are two major therapeutic areas in which even a chain of translatability from in vitro assays to preclinical models is often difficult to establish directly, as efficacy is a measure of a complex response at the tissue or the organism level. For example, late-stage cancers are frequently highly heterogeneous and adaptively independent of the known initiating oncogenic mutants, and neuropsychological disorders are often completely lacking in validated preclinical in vivo models. Here, it is much more difficult to correlate clinical outcomes to a single molecular target (target validation), cellular readout (in vitro model validation) or animal model (in vivo model validation) in the absence of additional studies.
Although these therapeutic areas are scientifically challenging and fraught with project-management risk, they include clinical indications that represent important unmet medical needs and business opportunities for the development of first-in-class therapeutics. To increase the probability of the predictive validity for models that lack a strong mechanistically defined chain of translatability, Vincent et al.8 proposed a set of guiding principles. The authors identified three key features (rule of 3) of the phenotypic assay — the assay system, stimulus and readout — that may enable the establishment of such a chain. First, the assay system must have a clear link to disease (for example, patient-derived primary cells or iPS-derived disease-relevant cell phenotypes) and aim to replicate relevant physiological aspects (for example, 3D growth or co-culture systems). Second, the assay readout should be as proximal as possible to the disease pathophysiology and clinical end point, ideally with a high degree of information. Third, the authors concluded that the stimuli used to induce a disease-like phenotype, which are often too simplistic, may result in the identification of stimulus- rather than disease-modifying compounds. Thus, systems that do not require an exogenous stimulus to induce the biological phenotype are preferred. Used as intended, these guidelines aim to increase both the biological space captured by the assay and the likelihood of the translation of the compounds and mechanism identified to patients. The excellent recent article by Horvath et al.22 provides further detailed descriptions and examples of systems that can enhance and validate the translational relevance of phenotypic assays.
Building the chain of translatability
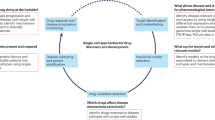
Disease understanding. Knowledge at the molecular level of the causes and drivers of the disease is a crucial success factor for PDD, as it is needed to select and to validate the best experimental cellular system and readouts to use (Fig. 1). Experience has shown that assays with generic readouts (for example, viability or apoptosis of cancer cell lines) are often not causally related to the disease biology pathways that we are attempting to modify, and thus are less likely to be useful in identifying novel and efficacious molecular MoAs.
a | The first step in establishing a chain of translatability for phenotypic drug discovery (PDD) is identifying a disease-associated molecular characteristic or signature (for example, a disease-associated gene expression profile, as shown, or the presence of a particular mutation in a protein) that differentiates the disease state (right) from normal physiology (left). When available human genetic or genomic data are insufficient to establish the causal components of this signature, animal models of the same disease can reconstruct earlier biological processes and associated pathway changes, providing a mechanistic bridge between alterations to normal physiology and the manifestations of the disease. b | Having identified the disease characteristics, cellular models aim to reconstruct a cellular phenotype that is as close as possible to the disease condition; for example, by incorporating a specific mutation in the cells or deriving cells from patients via induced pluripotent stem (iPS) cell generation. Specific disease-relevant stimuli may be required to model the cellular phenotype that is seen in the disease state. The mechanistic similarity of the model to the clinical disease is determined by a comparison of the molecular phenotype signatures. If the signatures are not sufficiently similar, the model is considered invalid. c | Phenotypic screening is conducted using a cellular model validated by the molecular phenotype. Prioritized hits from the primary screen may reveal different molecular phenotypes corresponding to different mechanisms of action (MoAs). Only MoAs that affect disease-relevant pathways will be evaluated for in vivo proof-of-concept. Nonspecific MoAs (represented by compound 3) can be eliminated using molecular phenotype information prior to advancing to in vivo proof-of-concept evaluation.
Although there remain diseases for which the molecular-mechanistic disease understanding may be insufficient to undertake effective PDD, this is changing rapidly as, for example, next-generation sequencing is contributing growing amounts of genetic data for many disorders23. For some diseases, especially those that require biopsies for diagnosis or treatment follow-up, there may be a large body of genetic and genomic information related to the baseline condition and to disease progression. This is, for instance, the case in kidney diseases24,25, for which this systems biology information has enabled the identification of new mechanisms and targets through the integration of large-scale genetic and molecular data with deep phenotypic information. This information has already been successfully translated into the identification of novel drugs through TDD approaches, such as the JAK2 inhibitor baricitinib26, which has reached late-stage clinical development for rheumatoid arthritis. However, attempts to use such knowledge to develop biologically informed PDD screens for kidney diseases have not yet been successful because it has not been possible to faithfully capture the complex human kidney pathophysiology in a suitable cellular assay system. Nevertheless, we are optimistic that these challenges can be overcome with the adoption of more sophisticated emerging models, such as organ-on-a-chip27.
Incomplete disease understanding is a limitation for the validation of phenotypic models and for hypothesis-driven molecular targets. This is illustrated by the challenges of transgenic mouse models of Alzheimer disease, which have been widely used, but have been unsuccessful in identifying clinically effective therapeutics so far. Although the molecular driver may be the same in both animal models and humans, the resulting pathogenic mechanism does not seem to be fully recapitulated in animal models28. Thus, capturing human disease relevance is the first step in the creation of a chain of translatability. Therefore, when starting a PDD programme, we need to be aware of the molecular mechanistic information that is available for the disease that we want to replicate in vitro and we should ideally place that molecular information in the context of clinical data. Are the translational data sufficient to identify well-validated molecular functional end points or predictive biomarkers for disease modification that can guide the PDD effort? If the same molecular functions and readouts can be encoded in a phenotypic screening model, then predictive validity is enhanced. Two key areas that can help to translate disease information into a screen with strong predictive validity are molecular phenotyping and advanced cellular models.
Molecular phenotyping. As we mentioned above, whereas knowledge of the molecular drivers of disease has been central to TDD approaches, the potential impact has not yet been fully realized in the context of PDD. A major challenge has been the substantial inability to translate such molecular MoA findings in humans in the context of a disease-relevant cell system that is appropriate for high-throughput hit identification. Molecular phenotyping29,30 — the ability to run high-throughput transcriptome analysis as a secondary or even a primary screen — thus holds promise as a technology to fully leverage this molecular information. Several such efforts are ongoing in the computational biology30,31,32, pharmaceutical33,34 and toxicogenomics35,36 fields. These efforts have the common aim of showing that the activity of signalling networks can be assessed based on a set of established key regulatory and effector genes. Molecular phenotype gene signatures have been shown by several groups31,33,36 to consistently deliver an accurate pathway-centric view of the biological system under study. The modulation of signalling networks identified in this way has been shown to be consistent with literature or experimental data assessed by different technologies.
In PDD, this means that we have a powerful tool to decode the effect of compounds on regulatory pathways in the context of the cellular model adopted. Pioneering work in this respect by Drawnel, Zhang and colleagues was recently published37. The authors were able to show, using a 917 human pathway reporter gene signature31, that molecular phenotyping can cluster compounds based on pathway profiles and can simultaneously dissect associations between pathway activities and disease phenotypes. Molecular phenotyping was applicable to compounds with a range of binding specificities and allowed false positives derived from high-content screening assays to be triaged. The approach was used to identify a class of calcium-signalling modulators that reversed disease-regulated pathways and phenotypes, which was validated by structurally distinct compounds in relevant classes37.
A similar approach was recently used to discover leptin sensitizers for diabetes. Here, researchers used profiling information on an active compound coupled with a search of the Broad Institute connectivity map (CMAP) to identify withaferin A as a novel leptin sensitizer38,39. We believe that such an approach, used in synergy with molecular disease information that is being generated by multiple initiatives40,41,42 that are sequencing DNA from patients with various diseases and phenotypically profiling them (for example, Genomics England41 and Genome Asia 100K43), may become a key enabler of future PDD. The critical value of this approach in establishing a chain of translatability is twofold. It first offers an unbiased diagnosis of the similarity between the disease state in humans and the molecular state of the discovery model (as shown in Fig. 2), and it also provides an evaluation of the extent to which a potential therapeutic modifies the molecular state towards the therapeutically desired state.
a | The disease model and the disease are represented as hidden state-dependent networks, and assay end points reflect the model state. On the left-hand side, a disease model that depends on a non-physiological external stimulus is shown, for which the assay end points are driven by network nodes that have no overlap with the disease state. Consequently, the assay end points are poorly predictive of a therapeutic effect. In the centre, the assay system state shares some common pathways with the disease, and the effects on some, but not all, of the assay end points may be predictive of a therapeutic effect. On the right-hand side, the network of the assay model strongly overlaps with the disease, and the effects on assay end points are highly predictive of a therapeutic effect. b | Illustration of how increasing the complexity of the assay model, from 2D cell culture (left) to a 3D culture (centre) and to a 3D co-culture system (right) might increase the overlap between the causal networks for the assay and disease end points, as well as the probability that a given assay readout will be predictive.
Advanced cellular models. The cellular screening system is a cornerstone of most successful attempts to identify potential novel drugs. Now, technological advances in cell and molecular biology are enabling the development of models that are likely to strengthen the chain of translatability even in model systems that have a reduced physiological complexity, by more closely modelling the disease-relevant cell or cells and tissue, and/or by focusing on the molecular and mechanistic phenotype.
In recent years, a broad arsenal of advanced cellular models44,45 have become available as microtechnologies have progressed: microprinted tumour spheroids46,47, 'tissue-on-a-chip' (Ref. 27), structured co-cultures and multicellular organoids48,49,50,51,52. Each system has its strengths and weaknesses reviewed in the references provided. Complementary advances in screening hardware, high-throughput cell assay technologies such as confocal high-content imaging systems and other methods for monitoring cell function are enabling more complex assays to be implemented for high-throughput lead discovery.
In addition to models that address the anatomical complexities of in vivo tissue structures, PDD efforts can leverage the predictive potential of iPS cell-based models53,54, which promise to replicate a disease in a dish (comprehensively reviewed by Avior et al.54). Models using iPS cells are especially powerful when studying high-penetrance monogenic disorders that are associated with a clear cellular phenotype. Breakthroughs in gene-editing technologies such as CRISPR–Cas are also likely to greatly increase the number and the diversity of genetically defined cellular models4,5, both conventional and iPS derived, used for PDD in the near future. At an even higher degree of complexity, an iPS cell-based model can be coupled to a molecular readout such as endogenous gene expression. For example, a screen carried out by Lee et al.55 assessed the ability of more than 6,000 molecules to restore the expression of IKK complex-associated protein (IKBAP), evaluated by real-time quantitative reverse-transcriptase PCR in cells derived from patients suffering from familial dysautonomia, who carry a hypomorphic mutation in the IKBAP gene. Nevertheless, even in the most advanced areas to make use of iPS-derived cell models — cardiac and neurological disorders — most studies have so far been aimed at validating the effects of existing drugs in a repurposing effort (see Table 3 in Avior et al.54) rather than identifying starting points for new drugs, reflecting the relatively short period for which iPS phenotypic screens have been pursued.
To address recurring concerns regarding the maturation state of iPS-derived cells, the use of patient-derived primary cells is a possible alternative. Sourcing such cells is still a rate-limiting step, but it is an area in which public–private partnerships and collaborations between biotech and pharma companies and disease foundations, as well as direct collaborations with health centres, hold great promise56. The translational value of these systems has been exemplified in the development of ivacaftor by scientists at Vertex Pharmceuticals57, including its label expansion based on an additional clinical trial of patients selected based on their genotype following compound testing against a wide range of genotypes encoded in patient-derived primary cells58. Fully differentiated patient-derived primary bronchial epithelial cells from healthy individuals or patients with cystic fibrosis harbouring the CFTRΔ508 mutation have also recently been used by scientists at Pfizer in a PDD project for cystic fibrosis59.
In the cancer drug discovery arena, results in the past few years from Cancer Genome Atlas data have highlighted the disconnections between even the best-characterized cancer cell line models and patients60,61,62. Concurrently, technical advances in generating patient-derived tumour models in vivo and in vitro are having radical effects on cancer drug research that ought to have a favourable impact on phenotypic discovery models. Two of the best-established patient-derived cancer cell culture systems are probably the glioma-derived neurosphere model and colorectal cancer-derived organoids. Both of these models, like the tumours from which they are derived, have a clear stem cell component that makes them amenable to genome editing63 and scalable for high-throughput screening64,65. The genetic diversity of tumours is thus not only captured but can also be made available through biobanks (for example, see Ref. 66).
However, powerful mechanistically informed cellular models do not always demand the use of a complex cellular system — the most important factor is that the molecular mechanism of the disease is reproduced in the observed phenotype of the discovery model (Fig. 2). In a striking example of a 'rule of 3 breaker', investigators from Roche and Novartis independently discovered small molecules that correct aberrant alternate splicing of the mRNA that codes for survival motor neuron 1 (SMN1), which is the root cause of the rare neurodegenerative disease, spinal muscular atrophy (SMA). Importantly, both molecules were optimized without prior knowledge of their molecular target by screening using generic cell lines that expressed reporter gene constructs designed to detect the alternative splicing of SMN1 (Refs 29,67). This was achieved despite a widely held consensus that reporter gene assays in engineered cell systems have low disease relevance and have arguably led to only a single marketed drug (vismodegib)6, presumably due to the smaller biological space that they probe (that is, direct transcriptional regulation) and high technical false-positive rates68. The cellular system in both cases was very simple but the proximity to the disease phenotype (that is, the ability to assess aberrant alternative splicing of a single gene) unparalleled. This resulted in an assay that was capable of guiding structure–activity relationship (SAR) studies, as well as producing molecules shown to be effective in disease models29,67, which are currently in late-stage clinical development for SMA.
Recent antibiotic discovery efforts have also provided instructive examples of the application of novel readout technologies to a simple 'classical' assay system to drive the exploration of new regions of chemical space. As discussed above, simple assays based on killing bacteria in vitro have a strong alignment to the desired pharmacodynamic effect in animal models and so are likely to identify hits that can be optimized into leads that show in vivo efficacy. The hurdle faced by antibiotic developers is that such chemical hits are frequently rediscoveries of known chemical scaffolds and thus are not appropriate for the development of drugs in novel classes, which are highly desirable in order to overcome resistance to antibiotics in existing classes. The challenge of identifying novel chemical starting points can be addressed by using discovery models that directly focus on a molecular phenotype. Two recent studies have illustrated this concept. Both groups took the approach of characterizing signatures predictive of the MoA of available antibiotics, and searched for novel hits that had a similar mechanistic profile but with an insufficient potency to be found in a classical functional antibacterial screen. Nonejuie et al.69 adopted high-content screening (HCS) to measure a large number of cellular features, whereas a second group at Roche extended this idea to the molecular level by using RNA-seq to identify drug-specific gene expression signatures (Zoffmann, S. et al., unpublished observations; presented at the 2016 Keystone symposium 'Modern Phenotypic Drug Discovery: Defining the Path Forward'). Both approaches indicated the ability to identify cellular pathways that predicted the MoAs of known antibacterial molecules. The team at Roche was able to further translate the results from RNA-seq to a higher-throughput bacteria reporter strain-based signature, and also reported that the signature allowed the identification of novel hits with the desired molecular MoA, thus providing novel chemical starting points.
To summarize, we believe that the ability to capture a disease-relevant molecular MoA in the screening system is a key enabling feature of PDD. Disease relevance can potentially be encoded in a simple cellular system, and novel chemical space may be explored by adopting innovative readouts for which a chain of translatability has been established. In the absence of the three core elements mentioned above — disease knowledge, replication and/or monitoring of the molecular MoA in vitro, and availability of a suitable cellular system8 — the probability of a successful PDD programme is greatly reduced.
Strategic considerations for PDD
A decision to pursue PDD versus TDD strategies requires a multitude of scientific, strategic and managerial factors to be considered. Below, we attempt to summarize the risks, costs and potential rewards of embarking on a PDD programme, based on our experience and that of others.
The promised payoffs of PDD include the identification of compounds that are more likely to translate to in vivo and clinical efficacy studies than TDD-derived compounds, or to be effective in a disease for which TDD cannot be applied (that is, a disease for which a validated molecular target is lacking), and/or the identification of compounds that act through novel mechanisms, potentially increasing the chances of differentiation from competitor compounds and existing standard treatments. Discovery of a novel target or molecular MoA provides an opportunity to develop a first-in-class medicine, which is a most desirable outcome for both patients and an industry facing regulatory and business headwinds for late entrants in an established target class.
PDD is not an easy path or a magic bullet, and there are sound reasons why TDD has been the dominant discovery paradigm of the past two decades. Without a high-confidence chain of translatability, the risk of clinical failure of PDD projects owing to a lack of efficacy is probably similar to that of a TDD project for a poorly validated target.
Building a rational, sustainable discovery pipeline around PDD presents considerable challenges. The early (preclinical) stages of PDD programmes — from lead-finding screen to clinical candidate — tend to require substantially greater resources than TDD. This is mostly due to the development of higher-complexity screening assays, and sometimes the concomitant use of small-molecule and genetic screens, as well as more challenging hit validation and target identification efforts for compound series of interest. However, the potential return on this greater investment stems from sampling a larger volume of potential target space, which may enable the discovery of either novel targets or unrecognized molecular MoAs. In light of this potential, it has been argued that the cost–benefit ratio for a single PDD project might be more comparable to several hypothesis-driven TDD projects for a given disorder70.
The perceived risk of advancing a compound with an incomplete understanding of its MoA or without identifying its molecular target varies, depending on the disease area (including the landscape of existing therapies), the existence of predictive and prognostic clinical biomarkers, safety concerns (see below) and the organizational strategy. In the absence of a target, the accumulation of mechanistic information (for example, identifying relevant signalling pathways or ruling out undesirable MoAs) can help to mitigate safety concerns. The most compelling argument in favour of advancing a PDD programme is the activity of the compound in a disease-relevant in vitro assay and animal disease model. As target identification is not always achievable71, the team and management must decide early on whether they are prepared to advance a drug candidate into preclinical or clinical development in the absence of knowledge of the identity of its target. The cost–benefit considerations of proceeding in the absence of target knowledge are very different depending on the indication, the strength of the chain of translatability, the presence of a causally related predictive biomarker, the unmet medical need, the competitive landscape and the risk-tolerance of the organization. However, the recurring myth that the definition of a molecular target is required for regulatory approval should be dispelled (see US Food and Drug Administration (FDA) guidance72): a substantial number of FDA-approved drugs (estimates range from 7%73 to 18%74) lack a defined molecular target.
It is generally agreed that the probability of technical success at the stage of the primary screen or of early discovery programmes is lower for PDD than for a typical TDD effort. Technical risks, which are discussed below, include challenging assay development, high false-positive hit rate, inability to establish SAR from the phenotypic assay, failure to generate a molecule that is suitable for in vivo proof- of-concept validation or the inability to identify the target of a compound series of interest. This reflects a front-loading of risk and a high bar to critical-path decision-making assay data early in the project. Moreover, because PDD emphasizes biological function it is likely that a PDD team will place high priority on early in vivo activity confirmation (with associated ADME (absorption, distribution, metabolism and excretion) and toxicology studies to deem the molecule safe) following the identification of compounds with very promising in vitro profiles.
Conversely, the most critical risk of the complementary TDD strategy for first-in-class drugs is target validation. Robust target validation is a serious challenge75,76, with poor linkage of a molecular target to the disease biology underlying a given indication estimated to contribute substantially to the ~50% of clinical failures that are caused by a lack of efficacy77.
Novelty of target and MoA is the second major potential advantage of PDD. In addition to identifying novel targets, PDD can contribute to improvements over existing therapies by identifying novel physiology for a known target, exploring 'undrugged' targets that belong to well known drug target classes or discovering novel MoAs, including new ways of interfering with difficult-to-drug targets. For example, the discovery of fingolimod78,79 led to the recognition of the role of sphingosine phosphate G protein-coupled receptors in lymphocyte egress from the thymus, and resulted in its approval as a first-in-class medication for multiple sclerosis80. Similarly, use of the well-validated maximal electroshock (MES) model for epilepsy enabled the discovery of lacosamide, which promotes the slow inactivation of voltage-gated sodium channels rather than directly blocking them81,82. More recently, a phenotypic screen for inhibitors of PCSK9 secretion delivered an unexpected molecular MoA in the form of a small molecule that selectively that stalls PCSK9 protein translation through an mRNA sequence-dependent interaction with the 80S ribosome17. Realistically, this molecular MoA would not have been pursued in a traditional TDD context and represents a clear example of how phenotypic strategies can expand our drug discovery horizon.
Taken together, these various considerations, including target validation, risk tolerance, breakthrough innovation versus incremental advancement, cost and competitive status, define a set of interconnected variables framing a complex decision-making landscape.
Operational aspects of PDD
Library selection. The strategic objectives of a PDD project should guide not only the selection of an appropriate screening model, as discussed and defined above, but also a decision on the chemical matter that is used for testing. Although not in the scope of this article, phenotypic screens of relatively small well-annotated tool compound collections are widely used to identify novel biology for molecular targets that belong to known drug target classes, and to explore drug-repurposing opportunities. Several recent articles have described the characterization and applications of such 'chemogenomics' libraries, with particular regard to understanding the specificity of the mechanism of action and the curation of nominal target identity83,84,85. The Chemical Probes Portal acts as a valuable public repository of such information.
By contrast, if compounds with novel targets are the goal, then diverse collections of novel compounds should be screened, given that an analysis by Santos et al.86 estimated that the molecular targets of known drugs and existing tool compounds constitute only 3% and 6% of the human proteome, respectively. Such compounds could be found in so-called 'dark chemical matter' (Ref. 87) — molecules that have been frequently screened in TDD projects but that have not shown any activity. Alternatively, natural products and their derivatives occupy an orthogonal chemical space to the majority of synthetic compounds and are frequently considered 'privileged', in that the structures of biologically relevant small molecules may be selected by evolution to engage protein-binding sites88 and thus may provide valuable chemical diversity for PDD. Natural products and their derivatives make up more than 25% of the new chemical entities that have been approved since 2010, an era in which natural products were not the focus of pharmaceutical research89. Interest in natural product screening is experiencing a renaissance (comprehensively reviewed by Harvey et al.90) that is contemporaneous with that of PDD and that is being driven by analogous reasons: technological advances and the depletion of the 'low-hanging fruit' in terms of validated and druggable target classes. A recent demonstration of the value of a natural-product-inspired diverse compound library as a substrate for phenotypic screening was the discovery of a novel anti-malarial compound that is efficacious in vivo, works on multiple parasite life stages and has a novel MoA91.
Although there are many important drug targets that do not require cell permeability, such as receptors, ion channels and secreted enzymes, a diverse phenotypic screening library requires a high probability of cellular permeability. Therefore, compounds with properties that are incompatible with this, such as peptidic compounds, higher molecular mass (>500–600 Da) and charged groups, although capable of providing optimizable lead matter in a biochemical TDD screen, should be deprioritized for inclusion. There are also growing efforts to define functional diversity using high-throughput approaches to determine bioactivity signatures. For example, scientists from the Broad Institute used a 'cell painting' method to profile a compound library and to prioritize compounds that displayed diverse bioactivity profiles92. Molecular phenotyping approaches, such as the Roche reporter gene31 and the National Institutes of Health/Broad Institute LINCS (Library of Integrated Network-based Cellular Signatures) L1000 panels93,94, are also potentially valuable in this regard. Given a library with a broad range of historical screening data, 'biological signatures' of compounds can be used to select for diversity, as well as to potentially aid in classifying hit MoAs85. However, prioritizing compounds for inclusion in a screening library based on previously observed biological effects is to some extent counter to the strategic goal of discovering molecules that have a selective effect on a disease-specific phenotype and model with no activity on 'normal' cellular physiology, so should not be applied too stringently. In fact, the exclusion of compounds based on promiscuity may be more important than the inclusion of nominally bioactive compounds, based on the demonstration that the absence of a history of hits in TDD projects (dark chemical matter) does not preclude future activity against a novel target87.
As well as being able to access the relevant compartment in the body, in most cases, a useful PDD screening hit needs to selectively bind to and modulate the function of a macromolecular target, whether it is an enzyme active site, an allosteric regulatory site, or an intra-molecular or an inter-molecular interaction. Therefore, it can be argued that the key factors in molecular features and properties of PDD screening collections compared with TDD screening collections are to place a premium on cellular permeability and to have sufficient structural complexity to confer some level of selectivity and low-micromolar affinity on molecular interactions. Although these requirements for selectivity and affinity usually argue against the utility of low-molecular-mass fragment compounds in a PDD screening library, there may be exceptions, such as the use of broadly targeted covalent chemical probes for the identification of targets of certain enzyme families, such as hydrolases and proteases95,96. A recent paper from the Cravatt laboratory also demonstrated that low-molecular-mass (~250 Da) fragment-like libraries can yield selective functionally active hits97. An important factor in making that particular fragment-based phenotypic screen useful and overcoming the inherent reduced binding specificity of smaller fragments was that each compound was also functionalized with a photoactivatable crosslinking moiety and an affinity tag, and thus any functional hits could be rapidly profiled for promiscuity of binding and target identity.
Regardless of the chemical diversity used in a PDD screen, the degree of compromise between throughput and assay complexity continues to be a challenge. In addition, screening a reference library of compounds with known targets and MoAs (that is, a chemogenomics library85) alongside a chemically diverse set is a stronglyrecommended step that will inform the design of an optimal hit triage strategy and will facilitate hit MoA and target deconvolution.
Hit triage. The selection of hits from phenotypic screens for further optimization is often considerably more complex than hit triaging from a target-based screen (Table 1). Hit rates for phenotypic screens of >1% are not uncommon (for example, see Refs 67,98), and thus hit lists of 1,000 to 10,000 or more compounds can be generated. Factors that contribute to the high confirmed hit rates for PDD include the larger molecular target space that is interrogated, and the potential to generate false positives through non-selective mechanisms. Therefore, rigorous follow-up assays to identify false positives are crucial. In addition to assays that monitor cytotoxicity, pleiotropic biological mechanisms that may mimic efficacy in the primary assay (for example, general secretion inhibition when monitoring levels of a secreted protein) must be anticipated and the appropriate counter screens are crucial. In our experience, primary hits from PDD are rarely target-specific hits. Following the confirmation of a PDD screening hit, it is thus important to establish SAR with a series of analogues of the same chemotype and to search for correlations, or lack thereof, with the desired (therapeutic) effect in order to distinguish target- or pathway-specific effects from undesired nonspecific effects. If a sufficient cluster of compounds for a specific chemotype is not present in the compound collection, a substantial investment in synthetic chemistry will potentially be required before a chemical series can be fully validated or invalidated.
In the case of TDD screens, structurally diverse hits often bind to the same site on the target, and thus shared pharmacophore features can increase hit confidence, and elements of different pharmacophores may eventually be combined. However, each chemotype arising from a phenotypic screen must be regarded as a standalone starting point, with a potentially distinct mode of action and target. Although this is a potential advantage in the long run, it greatly increases the complexity of the early stages of a project. Not every mode of action is suited for therapeutic intervention, as some may evoke the desired therapeutic effect while causing unacceptable side effects that are related to the same mechanism. In addition, it may be important to distinguish hits with a single mode of action from those with polypharmacology, in which the therapeutic effect is based on the synergistic or the additive interaction with multiple targets and pathways99. Although drugs based on polypharmacology can be effective6, such hits may not offer a tractable starting point for a target-agnostic lead optimization effort.
Prior knowledge of the activities and the target classes of chemotypes identified as hits might help point to targets or target pathways and may help to prioritize the hit list. In this way, the use of well-annotated compound collections85 for screening may simplify hit validation and prioritization. However, as previously mentioned, a reliance on such compound collections will probably bias the phenotypic screen towards known biology and targets. Alternatively, unbiased high-content and multi-parameter readouts of either the phenotypic screen itself or of the subsequent follow-up assays (for example, morphological, gene signature and proteomic changes) can greatly facilitate the clustering and evaluation of hits100,101,102. However, even with all readout and analytical tools in place, hit validation and the identification of relevant signalling pathways and molecular targets of phenotypic screening hits is often a lengthy and complex endeavour. Instructive examples for successful hit list triaging are the discovery of an inhibitor of the HCV NS5A protein15 and Porcupine inhibitors modulating oncogenic WNT pathway signalling103. In both cases, the primary hits were tested for specificity towards the inhibition of related biological systems — the replication of other flaviviruses and non-flaviviruses and Hedgehog pathway stimulation, respectively — as well as for overt cytotoxicity.
In our experience, if hits that only partially match the desired phenotypic profile are not stringently de-selected, then they can absorb the majority of resources, but very rarely end up being productive. A key lesson over the past few years has been that the follow-up of such suboptimal hits should be avoided, even if it means stopping the project in the absence of better matching hits. Another important success factor is the availability of a translational in vivo model to enable the accelerated testing of prioritized PDD-derived lead structures for, first, validating the pre-defined in vitro profile, second, enabling the final selection of chemotypes for further drug discovery investment, and third, allowing their efficient optimization.
Compound optimization. In some cases, phenotypic screening hits may provide access to several efficacious mechanisms. This allows project teams to evaluate multiple therapeutic mechanisms with different efficacy and safety profiles, to select the best fit for a desired indication (Fig. 3). So, there is the distinct possibility that a team may be optimizing several different chemical series with different targets.
Phenotypic drug discovery approaches will sample a wider range of potential therapeutic mechanisms than target-based approaches. With each mechanism possessing a unique efficacy versus safety profile, phenotypic screening offers an opportunity to identify and to evaluate multiple therapeutic options, including those based on previously unknown biology. Thus, for a target-based screen (left), the on-target safety liabilities of the lead compounds (usually evaluated in separate preclinical safety assays) will probably be the same for all lead compounds, as will the magnitude of their effect in a disease model. By contrast, a hypothetical phenotypic screen (right) yields compounds acting through three different targets, and for one of those targets, there are two distinct molecular mechanisms of action (MMoAs), one of which is the known target–MMoA combination shown for the target-based screen on the left-hand side. Each different target has its own distinct balance of efficacy and intrinsic safety liabilities.
One important risk when expanding and optimizing a compound series in the absence of a direct target engagement assay (whether biochemical or cellular) is that new analogues may alter their specificity profile or even act through a different target or mechanism from that of the original hit. Therefore, the application of data-rich mechanistic profiling assays, such as a transcriptional molecular phenotype, is as important for lead optimization as it is for hit triage.
The development of SARs and their use to drive improvement in a compound series is also considerably more complex for phenotypic assays, which are at least cell-based, if not tissue-based or whole-organism-based. As a result, additional variables must be accounted for during compound design and SAR data interpretation, such as cellular permeability, metabolic stability, potential polypharmacology and nonspecific binding to serum proteins. Although highly disease-relevant assays are often more difficult to prosecute, it is crucial that they deliver robust and reproducible data in order to successfully enable SAR determination (for example, a useful rule of thumb is that an assay must reproducibly detect a threefold difference in compound potency to be useful). However, it is important to remember that medicinal chemists have proved capable of delivering clinical candidates in the absence of target binding assays and structure-based design81,104.
Three key parameters have been proposed to be essential to confidently test a mechanistic hypothesis in the clinic: compound exposure at the site of action, target binding and expression of functional pharmacological activity105. As target engagement information may be out of reach for a PDD programme, the third criterion will have an increased importance in ensuring that the therapeutic hypothesis is effectively tested in humans. Biomarkers of either compound pharmacology or of the disease itself will thus be crucial for programme progression70. It is important to note that, as ex vivo assays using patient-derived cells and tissues become more prevalent, the need for animal models of disease to provide confidence in translation to patients may become less acute. This may represent a considerable benefit for those disorders with poorly predictive models2,106. However, the utility of testing compounds in other species for safety, pharmacokinetic and pharmacodynamic purposes will remain.
Safety lessons. As PDD inherently identifies compounds with unknown targets, there are accompanying risks of hits engaging a target or of having an MoA with significant safety liabilities. The safety de-risking of target-based drug discovery programmes is based on multiple types of information available to researchers, as well as unbiased in vitro and in vivo experimental safety studies (Box 1). Target-related knowledge, including known biological roles, expression pattern, target-deficient or mutant organisms and the existence of closely related proteins, constitutes a substantial portion of this information. If a target or MoA can be directly identified during a phenotypic screen by using an annotated small-molecule library or a genetic screening approach, the resulting hits can be de-risked through this general approach. However, phenotypic screens will usually identify compounds with unknown mechanisms, necessitating a different set of safety considerations.
An additional, underappreciated distinction exists between the efficacious mechanisms that are obtained through a phenotypic approach and those derived from known targets in terms of safety prospects. For target-based programmes, scientists can first evaluate whether the prospective balance of the efficacy and the safety potential is attractive enough to warrant inclusion in a project portfolio. Although not fool-proof, this target-based evaluation is likely to remove many mechanisms with poor safety prognostics from consideration. Without this level of review, it can be expected that a greater proportion of phenotypic screening-derived mechanisms, although efficacious, will have significant safety liabilities (for example, target B in Fig. 3).
Two potential issues may further complicate safety de-risking of a compound series in the absence of a molecular target. First, target biology may substantially differ between rodents and humans. An in vivo safety study in a second species, as required by the FDA before human testing, can help to address this concern. Second, the lack of target information leads to a diminished ability to design bespoke experiments that address specific safety issues early on (for example, detailed cardiovascular studies for targets that are expressed in heart tissue or the analysis of infection risk for targets that are important for the adaptive or innate immune system). Overall, the main added risk in the absence of a target hypothesis is the delayed identification of an unacceptable safety profile and the resulting loss of the investment made thus far in a given compound series.
Accordingly, phenotypic projects may benefit from earlier and larger investment in safety experiments compared with target-based programmes. Most, but probably not all, of these undesirable mechanisms can be rapidly eliminated during the hit triage through the thoughtful use of biological counter-screens. Additionally, many of the technological advances previously described for assay development also enable predictive toxicology. These include the application of transcriptomic and proteomic profiling, and the use of iPSC-derived models for cardiotoxicity107 and hepatoxicity108 assays. Finally, compound-derived versus mechanism-related toxicity can still potentially be resolved by conducting experiments with pairs of closely related active and inactive compounds81, similar to what is done for target validation109. Another strategy is, of course, to identify the target before large-scale preclinical investment or clinical entry.
After the screen: is target identification essential? PDD and target deconvolution are inextricably intertwined, both in the strategic considerations of whether to pursue PDD, and in its practice. Advances in target identification (TID) technologies, which have been extensively reviewed elsewhere110–115 and which are likely to be further advanced by genomics approaches116, may decrease the target identification hurdle and change the risk equation for PDD. However, the resource costs of TID and the risk of failing to identify the causal molecular target are still major potential roadblocks, depending on the discovery strategy.
At a panel discussion on TID and associated risks at the 2016 Keystone symposium 'Modern Phenotypic Drug Discovery: Defining the Path Forward', in which both biotech and large pharmaceutical companies were represented, there was a general agreement from both groups that the absence of target knowledge is not necessarily a no-go gate for the hit-to-lead activities, as long as SARs are built on a robust phenotypic assay and a highly relevant readout. However, the discussion did reveal some key institutional differences. While for some companies, TID is a mandatory requirement before investing substantial chemistry resources and advancing to lead identification, other companies are prepared to conduct SAR and TID efforts in parallel, with a view to obtaining the clinical candidate, as well as the molecular MoA, before clinical studies. An outstanding example of this mindset is the development by Novartis of a drug for SMA that corrects an SMN1 pre-mRNA splicing defect (mentioned above), for which there was a willingness to initiate human studies even in the absence of a known target (although the MoA was eventually uncovered before the initiation of clinical trials)29. In this instance, the risk tolerance may be attributable to the combination of an acute unmet medical need coupled with high confidence in the translatability of the molecular MoA to clinical efficacy in a genetically defined patient population. In other words, all the key elements of the chain of translatability were in place.
In general, the consensus from the Keystone panel discussion was that smaller biotech companies are prepared to progress towards clinical trials without TID, while following preclinical regulatory safety guidelines. TID is more likely to be perceived by those in biotech companies as diverting limited resources away from the delivery of a clinical asset for which the degree of resource investment needed to gain knowledge that is actionable in terms of clinical development can be daunting, with an uncertain outcome70. Conversely, most large pharmaceutical companies strongly emphasize TID as a crucial component of project progression and prioritization as part of a portfolio that also contains targeted drugs. In this business environment, the failure to assign an MoA and/or a target to a new molecular entity (NME) is frequently considered a major risk factor for clinical development and regulatory approval, including the safety challenges discussed above.
The decisions around TID also have strategic relevance in the context of the competitive environment. For a disease area with several NMEs in development or with an already available standard-of-care treatment, can the patients who will maximally benefit from the NME be identified in the absence of a mechanism and, therefore, a proximal PD marker? Once the NME is approved, in the context of life cycle management, how can the market be expanded towards new indications in the absence of a target or MoA? Although there are examples of the empirical discovery of new therapeutic uses of drugs (for example, topiramate117 was initially approved for epilepsy and was later approved for migraine prophylaxis118), this is a valid concern. As highlighted above, TID may have a greater impact in the context of clinical development than in the early drug discovery process. This would help to explain why successful TID is more likely to be considered important by large pharmaceutical companies carrying out phase II/III studies towards regulatory approval than smaller biotech companies that are more usually focused on reaching a phase I/II milestone.
Another key value of TID to be considered is whether it can prompt the inception of a TDD programme. For instance, the anti-epileptic drug levetiracetam, which was identified in a target-agnostic model (audiogenic seizure-susceptible mice119) was the precursor to several rationally designed follow-on drugs119. These follow-on drugs were discovered following an impressive TID effort conducted by scientists from UCB Pharma, who unravelled the ubiquitous synaptic vesicle glycoprotein SV2A as the target of levetiracetam120.
Overall conclusions
Rather than being viewed as opposing alternatives in novel drug discovery, PDD and TDD should be seen as complementary approaches that can together increase the odds of discovering and developing drugs with novel efficacious molecular MoAs. The unique promise of PDD is its ability to exploit a disease phenotype to discover novel treatments for diseases for which the root cause is unknown, complex or multifactorial, and for which scientific understanding is insufficient to provide valid molecular targets. However, PDD should not be regarded simply as an alternative screening technology or as an easy fix to the challenges of clinical attrition rates or R&D productivity. We referred above to PDD as being at risk of undergoing a hype cycle. It is our intention to constructively minimize overly optimistic expectations for 'quick wins' from PDD, but also to provide advice and encouragement to ameliorate the potential for a trough of disillusionment that may arise when organizations are not frequently rewarded with first-in-class or best-in-class drugs or even with tractable leads from phenotypic screens.
We have reviewed the relevance of the chain of translatability, a continuum that links the human disease biology at one end, through the phenotypic system used for compound screening, to therapeutic activity. Enabling tools and technologies for rational, mechanistically informed phenotypic assays are rapidly advancing at all levels: genetically defined cellular models of disease, complex cellular systems mirroring the in vivo state, ever-more informative high-content imaging technologies and sophisticated sequencing-based methods to investigate disease relevance in multi-well plate assays. We have also highlighted that incorporating the precise molecular MoA — even in a simple cell system, as in the case of SMA mentioned above — is at the heart of any successful PDD approach. In summary, we believe that the ability to capture disease-relevant regulatory pathways in the screening system is a key enabling feature of PDD, and that exploration of chemical space can be carried out through the adoption of innovative readouts, as long as a chain of translatability exists.
Rare diseases may represent a 'sweet spot' for the application of PDD in academic settings. In addition to a strong chain of translatability, other elements that are important for success are in place: access to funding provided by governments, charities and patient associations121, deep disease molecular knowledge produced by highly engaged physician-scientist-led groups, and the ability to generate iPS-derived and/or gene-edited cell models that can be readily screened in publicly funded high-quality screening centres. We have also tried to convey our experiences and perspectives of the challenges of integrating PDD into organizations with a strong target-centric perspective and history. From the outset, managers and scientists need to understand and accept the different risk/benefit considerations associated with PDD compared with classical target-based approaches. PDD is highly likely to involve greater early-stage resource requirements, greater uncertainty and to entail longer timelines than TDD. Furthermore, the critical path for project progression and the criteria for validation of chemical matter are more fluid and empirical. Decision gates are likely to be different: relevance to the disease mechanism is crucial, potency is secondary and advancing to in vivo proof-of-concept efficacy studies is a crucial go/no-go gate to be reached as soon as possible. Based on the input of many practitioners who have shared their experiences, we recommend only undertaking a PDD effort if the disease-relevant molecular MoA is well understood and/or when it is possible to sufficiently establish a chain of translatability in a screening model. We surmise that not meeting these conditions is the main reason for the failure of PDD projects. Similarly, we suggest that if a hit is non-selective in terms of the desired phenotypic profile, it almost certainly cannot be optimized to be selective — an important difference from most TDD programmes.
A crucial strategic consideration is how strongly to tie project advancement, and perhaps even clinical development, to TID. Novelty, unmet medical need, compelling in vivo biology and expected safety margin have triggered clinical decisions in the absence of a precise molecular target or molecular MoA for drugs that are currently on the market or in late-stage clinical trials, including pirfenidone122, its direct competitor nintedanib123, and thalidomide and its analogues lenalidomide and pomalidomide. Guidance from such examples can inform decisions about whether and when to spend resources for target identification and detailed molecular MoA studies.
PDD is a challenging drug discovery strategy on multiple levels, but it has a successful track record of delivering first-in-class drugs. It is a powerful approach to exploit the novel biological space of undrugged or unknown targets and poorly understood disease mechanisms, providing a route to enhance innovation in the pharmaceutical industry and to deliver truly novel therapeutics for unmet medical needs.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Swinney, D. C. & Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 10, 507–519 (2011).
Scannell, J. W. & Bosley, J. When quality beats quantity: decision theory, drug discovery, and the reproducibility crisis. PLoS ONE 11, e0147215 (2016).
Wagner, B. K. & Schreiber, S. L. The power of sophisticated phenotypic screening and modern mechanism-of-action methods. Cell Chem. Biol. 23, 3–9 (2016).
Shi, Y., Inoue, H., Wu, J. C. & Yamanaka, S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 16, 115–130 (2017).
Fellmann, C., Gowen, B. G., Lin, P. C., Doudna, J. A. & Corn, J. E. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 16, 89–100 (2017).
Eder, J., Sedrani, R. & Wiesmann, C. The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug Discov. 13, 577–587 (2014).
Moffat, J. G., Rudolph, J. & Bailey, D. Phenotypic screening in cancer drug discovery — past, present and future. Nat. Rev. Drug Discov. 13, 588–602 (2014).
Vincent, F. et al. Developing predictive assays: the phenotypic screening “rule of 3”. Sci. Transl Med. 7, 293ps15 (2015).
Mullard, A. The phenotypic screening pendulum swings. Nat. Rev. Drug Discov. 14, 807–809 (2015).
Lee, J. A. & Berg, E. L. Neoclassic drug discovery: the case for lead generation using phenotypic and functional approaches. J. Biomol. Screen. 18, 1143–1155 (2013).
Gonzalez-Munoz, A. L., Minter, R. R. & Rust, S. J. Phenotypic screening: the future of antibody discovery. Drug Discov. Today 21, 150–156 (2016).
Andries, K. et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227 (2005).
Ma, H. et al. Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor beta-D-2′-Deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) and identification of a novel active 5′-triphosphate species. J. Biol. Chem. 282, 29812–29820 (2007).
Queiroz, E. F., Wolfender, J. L. & Hostettmann, K. Modern approaches in the search for new lead antiparasitic compounds from higher plants. Curr. Drug Targets 10, 202–211 (2009).
Gao, M. et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465, 96–100 (2010).
Burke, A. C., Dron, J. S., Hegele, R. A. & Huff, M. W. PCSK9: regulation and target for drug development for dyslipidemia. Annu. Rev. Pharmacol. Toxicol. 57, 223–244 (2016).
Petersen, D. N. et al. A small-molecule anti-secretagogue of PCSK9 targets the 80S ribosome to inhibit PCSK9 protein translation. Cell Chem. Biol. 23, 1362–1371 (2016).
Swinney, D. C. & Xia, S. The discovery of medicines for rare diseases. Future Med. Chem. 6, 987–1002 (2014).
Swinney, D. C. Challenges and hurdles to business as usual in drug development for treatment of rare diseases. Clin. Pharmacol. Ther. 100, 339–341 (2016).
Nystrom, A. et al. Losartan ameliorates dystrophic epidermolysis bullosa and uncovers new disease mechanisms. EMBO Mol. Med. 7, 1211–1228 (2015).
Fishman, M. C. Power of rare diseases: found in translation. Sci. Transl Med. 5, 201ps11 (2013).
Horvath, P. et al. Screening out irrelevant cell-based models of disease. Nat. Rev. Drug Discov. 15, 751–769 (2016).
Alfoldi, J. & Lindblad-Toh, K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 23, 1063–1068 (2013).
Mariani, L. H., Pendergraft, W. F. III & Kretzler, M. Defining glomerular disease in mechanistic terms: implementing an integrative biology approach in nephrology. Clin. J. Am. Soc. Nephrol. 11, 2054–2060 (2016).
Ju, W. et al. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci. Transl Med. 7, 316ra193 (2015).
Brosius, F. C., Tuttle, K. R. & Kretzler, M. JAK inhibition in the treatment of diabetic kidney disease. Diabetologia 59, 1624–1627 (2016).
Wang, L. et al. A disease model of diabetic nephropathy in a glomerulus-on-a-chip microdevice. Lab Chip 17, 1749–1760 (2017).
Keene, C. D. et al. Neuropathological assessment and validation of mouse models for Alzheimer's disease: applying NIA-AA guidelines. Pathobiol. Aging Age Relat. Dis. 6, 32397 (2016).
Palacino, J. et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat. Chem. Biol. 11, 511–517 (2015).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Zhang, J. D., Kung, E., Boess, F., Certa, U. & Ebeling, M. Pathway reporter genes define molecular phenotypes of human cells. BMC Genomics 16, 342 (2015).
Zhang, J. D., Schindler, T., Kung, E., Ebeling, M. & Certa, U. Highly sensitive amplicon-based transcript quantification by semiconductor sequencing. BMC Genomics 15, 565 (2014).
Zhang, J. D., Berntenis, N., Roth, A. & Ebeling, M. Data mining reveals a network of early-response genes as a consensus signature of drug-induced in vitro and in vivo toxicity. Pharmacogenomics J. 14, 208–216 (2014).
Moisan, A. et al. White-to-brown metabolic conversion of human adipocytes by JAK inhibition. Nat. Cell Biol. 17, 57–67 (2015).
Nuwaysir, E. F., Bittner, M., Trent, J., Barrett, J. C. & Afshari, C. A. Microarrays and toxicology: the advent of toxicogenomics. Mol. Carcinogen. 24, 153–159 (1999).
Merrick, B. A., Paules, R. S. & Tice, R. R. Intersection of toxicogenomics and high throughput screening in the Tox21 program: an NIEHS perspective. Int. J. Biotechnol. 14, 7–27 (2015).
Drawnel, F. M. et al. Molecular phenotyping combines molecular information, biological relevance, and patient data to improve productivity of early drug discovery. Cell Chem. Biol. 24, 624–634.e3 (2017).
Lee, J. et al. Withaferin A is a leptin sensitizer with strong antidiabetic properties in mice. Nat. Med. 22, 1023–1032 (2016).
Liu, J., Lee, J., Salazar Hernandez, M. A., Mazitschek, R. & Ozcan, U. Treatment of obesity with celastrol. Cell 161, 999–1011 (2015).
Amos, C. I. et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomarkers Prev. 26, 126–135 (2017).
Marx, V. The DNA of a nation. Nature 524, 503–505 (2015).
Chambers, J. C. et al. The South Asian genome. PLoS ONE 9, e102645 (2014).
Cyranoski, D. Genomics takes hold in Asia. Nature 456, 12 (2008).
Nath, S. & Devi, G. R. Three-dimensional culture systems in cancer research: focus on tumor spheroid model. Pharmacol. Ther. 163, 94–108 (2016).
Esch, E. W., Bahinski, A. & Huh, D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 14, 248–260 (2015).
Ham, S. L., Atefi, E., Fyffe, D. & Tavana, H. Robotic production of cancer cell spheroids with an aqueous two-phase system for drug testing. J. Vis. Exp. 23, e52754 (2015).
Thakuri, P. S., Ham, S. L., Luker, G. D. & Tavana, H. Multi-parametric analysis of oncology drug screening with aqueous two-phase tumor spheroids. Mol. Pharm. 13, 3724–3735 (2016).
Friedrich, J., Seidel, C., Ebner, R. & Kunz-Schughart, L. A. Spheroid-based drug screen: considerations and practical approach. Nat. Protoc. 4, 309–324 (2009).
Katt, M. E., Placone, A. L., Wong, A. D., Xu, Z. S. & Searson, P. C. In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 4, 12 (2016).
Gunter, J. et al. Microtissues in cardiovascular medicine: regenerative potential based on a 3D microenvironment. Stem Cells Int. 2016, 9098523 (2016).
Weiswald, L. B., Bellet, D. & Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 17, 1–15 (2015).
Hild, M. & Jaffe, A. B. Production of 3D airway organoids from primary human airway basal cells and their use in high-throughput screening. Curr. Protoc. Stem Cell Biol. 37, IE.9.1–IE.9.15 (2016).
Heilker, R., Traub, S., Reinhardt, P., Scholer, H. R. & Sterneckert, J. iPS cell derived neuronal cells for drug discovery. Trends Pharmacol. Sci. 35, 510–519 (2014).
Avior, Y., Sagi, I. & Benvenisty, N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 17, 170–182 (2016).
Lee, G. et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat. Biotechnol. 30, 1244–1248 (2012).
Edwards, A. M. et al. Preclinical target validation using patient-derived cells. Nat. Rev. Drug Discov. 14, 149–150 (2015).
Van Goor, F. et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl Acad. Sci. USA 106, 18825–18830 (2009).
Yu, H. et al. Ivacaftor potentiation of multiple CFTR channels with gating mutations. J. Cyst. Fibros. 11, 237–245 (2012).
Berg, A. et al. A phenotypic screen for corrector discovery using a surface liquid readout in F508del primary airway epithelia. Pediatr. Pulmonol. 50, abstr. 181 (2015).
Klijn, C. et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33, 306–312 (2015).
Goodspeed, A., Heiser, L. M., Gray, J. W. & Costello, J. C. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol. Cancer Res. 14, 3–13 (2016).
Cascorbi, I. & Werk, A. N. Advances and challenges in hereditary cancer pharmacogenetics. Expert Opin. Drug Metab. Toxicol. 13, 73–82 (2017).
O'Duibhir, E., Carragher, N. O. & Pollard, S. M. Accelerating glioblastoma drug discovery: convergence of patient-derived models, genome editing and phenotypic screening. Mol. Cell. Neurosci. 80, 198–207 (2017).
Quartararo, C. E., Reznik, E., deCarvalho, A. C., Mikkelsen, T. & Stockwell, B. R. High-throughput screening of patient-derived cultures reveals potential for precision medicine in glioblastoma. ACS Med. Chem. Lett. 6, 948–952 (2015).
Verissimo, C. S. et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife 5, e18489 (2016).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Naryshkin, N. A. et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 345, 688–693 (2014).
Auld, D. S. et al. Characterization of chemical libraries for luciferase inhibitory activity. J. Med. Chem. 51, 2372–2386 (2008).
Nonejuie, P., Burkart, M., Pogliano, K. & Pogliano, J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl Acad. Sci. USA 110, 16169–16174 (2013).
Swinney, D. C. The value of translational biomarkers to phenotypic assays. Front. Pharmacol. 5, 171 (2014).
Klotz, J. Phenotypic screening, take two. SciBX http://dx.doi.org/10.1038/scibx.2012.380 (2012).
Center for Drug Evaluation and Research (CDER) & Center for Biologics Evaluation and Research (CBER). Guidance for industry content and format of investigational new drug applications (INDs) for phase 1 studies of drugs, including well-characterized, therapeutic, biotechnology-derived products. FDA http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm071597.pdf (1995).
Drews, J. Drug discovery: a historical perspective. Science 287, 1960–1964 (2000).
Overington, J. P., Al-Lazikani, B. & Hopkins, A. L. How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 (2006).
Begley, C. G. & Ellis, L. M. Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
Prinz, F., Schlange, T. & Asadullah, K. Believe it or not: how much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov. 10, 712 (2011).
Hay, M., Thomas, D. W., Craighead, J. L., Economides, C. & Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 32, 40–51 (2014).
Kiuchi, M. et al. Synthesis and immunosuppressive activity of 2-substituted 2-aminopropane-1,3-diols and 2-aminoethanols. J. Med. Chem. 43, 2946–2961 (2000).
Kovarik, J. M., Schmouder, R. L. & Slade, A. J. Overview of FTY720 clinical pharmacokinetics and pharmacology. Ther. Drug Monit. 26, 585–587 (2004).
Brinkmann, V. et al. Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 9, 883–897 (2010).
Choi, D., Stables, J. P. & Kohn, H. Synthesis and anticonvulsant activities of N-benzyl-2-acetamidopropionamide derivatives. J. Med. Chem. 39, 1907–1916 (1996).
Errington, A. C., Stohr, T., Heers, C. & Lees, G. The investigational anticonvulsant lacosamide selectively enhances slow inactivation of voltage-gated sodium channels. Mol. Pharmacol. 73, 157–169 (2008).
Arrowsmith, C. H. et al. The promise and peril of chemical probes. Nat. Chem. Biol. 11, 536–541 (2015).
Wang, Y. et al. Evidence-based and quantitative prioritization of tool compounds in phenotypic drug discovery. Cell Chem. Biol. 23, 862–874 (2016).
Jones, L. H. & Bunnage, M. E. Applications of chemogenomic library screening in drug discovery. Nat. Rev. Drug Discov. 16, 285–296 (2017).
Santos, R. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017).
Wassermann, A. M. et al. Dark chemical matter as a promising starting point for drug lead discovery. Nat. Chem. Biol. 11, 958–966 (2015).
Wetzel, S., Bon, R. S., Kumar, K. & Waldmann, H. Biology-oriented synthesis. Angew Chem. Int. Ed. 50, 10800–10826 (2011).
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016).
Harvey, A. L., Edrada-Ebel, R. & Quinn, R. J. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 14, 111–129 (2015).
Kato, N. et al. Diversity-oriented synthesis yields novel multistage antimalarial inhibitors. Nature 538, 344–349 (2016).
Wawer, M. J. et al. Toward performance-diverse small-molecule libraries for cell-based phenotypic screening using multiplexed high-dimensional profiling. Proc. Natl Acad. Sci. USA 111, 10911–10916 (2014).
De Wolf, H., De Bondt, A., Turner, H. & Gohlmann, H. W. Transcriptional characterization of compounds: lessons learned from the public LINCS data. Assay Drug Dev. Technol. 14, 252–260 (2016).
Liu, C. et al. Compound signature detection on LINCS L1000 big data. Mol. Biosyst. 11, 714–722 (2015).
Matthews, M. L. et al. Chemoproteomic profiling and discovery of protein electrophiles in human cells. Nat. Chem. 9, 234–243 (2017).
Backus, K. M. et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016).
Parker, C. G. et al. Ligand and target discovery by fragment-based screening in human cells. Cell 168, 527–541.e29 (2017).
Rottmann, M. et al. Spiroindolones, a potent compound class for the treatment of malaria. Science 329, 1175–1180 (2010).
Zhang, W., Bai, Y., Wang, Y. & Xiao, W. Polypharmacology in drug discovery: a review from systems pharmacology perspective. Curr. Pharm. Des. 22, 3171–3181 (2016).
Taylor, D. L. Past, present, and future of high content screening and the field of cellomics. Methods Mol. Biol. 356, 3–18 (2007).
Perlman, Z. E. et al. Multidimensional drug profiling by automated microscopy. Science 306, 1194–1198 (2004).
Caie, P. D. et al. High-content phenotypic profiling of drug response signatures across distinct cancer cells. Mol. Cancer Ther. 9, 1913–1926 (2010).
Liu, J. et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl Acad. Sci. USA 110, 20224–20229 (2013).
Black, J. Nobel lecture in physiology or medicine — 1988. Drugs from emasculated hormones: the principle of syntopic antagonism. In Vitro Cell Dev. Biol. 25, 311–320 (1989).
Morgan, P. et al. Can the flow of medicines be improved? Fundamental pharmacokinetic and pharmacological principles toward improving phase II survival. Drug Discov. Today 17, 419–424 (2012).
van der Worp, H. B. et al. Can animal models of disease reliably inform human studies? PLoS Med. 7, e1000245 (2010).
Sirenko, O. et al. In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-derived model. Toxicol. Appl. Pharmacol. 322, 60–74 (2017).
Mann, D. A. Human induced pluripotent stem cell-derived hepatocytes for toxicology testing. Expert Opin. Drug Metab. Toxicol. 11, 1–5 (2015).
Sweis, R. F. Target (in)validation: a critical, sometimes unheralded, role of modern medicinal chemistry. ACS Med. Chem. Lett. 6, 618–621 (2015).
Saxena, C. Identification of protein binding partners of small molecules using label-free methods. Expert Opin. Drug Discov. 11, 1017–1025 (2016).
Fetz, V., Prochnow, H., Bronstrup, M. & Sasse, F. Target identification by image analysis. Nat. Prod. Rep. 33, 655–667 (2016).
Schirle, M. & Jenkins, J. L. Identifying compound efficacy targets in phenotypic drug discovery. Drug Discov. Today 21, 82–89 (2016).
Adams, R., Steckel, M. & Nicke, B. Functional genomics in pharmaceutical drug discovery. Handb. Exp. Pharmacol. 232, 25–41 (2016).
Moore, J. D. The impact of CRISPR–Cas9 on target identification and validation. Drug Discov. Today 20, 450–457 (2015).
Lee, H. & Lee, J. W. Target identification for biologically active small molecules using chemical biology approaches. Arch. Pharmacol. Res. 39, 1193–1201 (2016).
Nijman, S. M. Functional genomics to uncover drug mechanism of action. Nat. Chem. Biol. 11, 942–948 (2015).
Maryanoff, B. E. Phenotypic assessment and the discovery of topiramate. ACS Med. Chem. Lett. 7, 662–665 (2016).
Silberstein, S. Topiramate in migraine prevention. Headache 45 Suppl. 1, S57–S65 (2005).
Rogawski, M. A. Brivaracetam: a rational drug discovery success story. Br. J. Pharmacol. 154, 1555–1557 (2008).
Kaminski, R. M. et al. SV2A protein is a broad-spectrum anticonvulsant target: functional correlation between protein binding and seizure protection in models of both partial and generalized epilepsy. Neuropharmacology 54, 715–720 (2008).
Mavris, M. & Le Cam, Y. Involvement of patient organisations in research and development of orphan drugs for rare diseases in europe. Mol. Syndromol. 3, 237–243 (2012).
Nakazato, H., Oku, H., Yamane, S., Tsuruta, Y. & Suzuki, R. A novel anti-fibrotic agent pirfenidone suppresses tumor necrosis factor-alpha at the translational level. Eur. J. Pharmacol. 446, 177–185 (2002).
Roth, G. J. et al. Nintedanib: from discovery to the clinic. J. Med. Chem. 58, 1053–1063 (2015).
Acknowledgements
The authors thank A. Subramanian and J. Rudolph for helpful discussions and comments and acknowledge the invaluable contributions of the participants of the 2015 New York Academy of Sciences Symposium and the 2016 Keystone Symposium on phenotypic drug discovery. The idea to gather and share experiences from multiple companies coalesced from conversations with many outstanding scientists during the breaks of those two landmark conferences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
John G. Moffat is an employee of Genentech. Fabien Vincent is an employee of Pfizer. Jonathan A. Lee is an employee of Eli Lilly. Jörg Eder is an employee of Novartis. Marco Prunotto is an employee of Roche.
Related links
DATABASES
Glossary
- Chain of translatability
-
A molecular-level association between the mechanisms that drive the assay phenotype, the preclinical disease model and the human disease.
- Molecular phenotype
-
Gene-level and pathway-level 'omics' signatures shared by disease model and disease state that correspond to and are predictive of disease state versus normal state.
- Organoids
-
In vitro 3D cellular clusters derived from primary tissue or stem cells that show similar characteristics to the tissue of origin; for example, beating cardiomyocytes.
- PCSK9
-
(Proprotein convertase subtilisin/kexin type 9). A secreted protein mainly expressed in the liver. Studies of naturally occurring human genetic variants in PCSK9 provided strong evidence that PCSK9 inhibitors could reduce plasma levels of low-density lipoprotein cholesterol and reduce cardiovascular risk.
- Pharmacophores
-
Groups of molecular features that mediate interactions between a compound and a particular biological target macromolecule and trigger (or block) its biological response.
- RNA-seq
-
Uses rapid sequencing technologies to identify the presence and quantity of RNAs in a biological sample at a given moment in time.
- Rule of 3
-
Three technical factors that influence the probability that a phenotypic assay will identify relevant molecules that affect relevant disease mechanisms: biological system, stimulus and readout.
Rights and permissions
About this article
Cite this article
Moffat, J., Vincent, F., Lee, J. et al. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat Rev Drug Discov 16, 531–543 (2017). https://doi.org/10.1038/nrd.2017.111
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2017.111
This article is cited by
-
A catalytic process enables efficient and programmable access to precisely altered indole alkaloid scaffolds
Nature Chemistry (2024)
-
Unexpected inhibition of the lipid kinase PIKfyve reveals an epistatic role for p38 MAPKs in endolysosomal fission and volume control
Cell Death & Disease (2024)
-
High-content screening identifies a small molecule that restores AP-4-dependent protein trafficking in neuronal models of AP-4-associated hereditary spastic paraplegia
Nature Communications (2024)
-
Predicting compound activity from phenotypic profiles and chemical structures
Nature Communications (2023)
-
Discovery of senolytics using machine learning
Nature Communications (2023)