Key Points
-
The recent significant advances in the development of single-cell analytical tools have enabled increasingly deep cellular analyses at the genomic, transcriptomic and proteomic levels.
-
Genomic and transcriptomic methods utilize next-generation sequencing technologies, which complement protocols for improved quantification and cost-effective analyses of statistically significant numbers of single cells.
-
Single-cell proteomics methods range from cytometry tools to microchip platforms. All these methods rely on antibodies, but different platforms yield different levels of quantification.
-
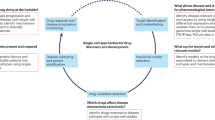
Single-cell analyses reveal biology that is masked when cell populations or tissues are analysed. Illustrative examples include tracing the lineage of diseased cells back to the healthy tissue of origin, or a deep analysis of how targeted inhibitors can alter the structure of signalling pathways.
-
Single-cell analysis tools are already playing important parts in drug discovery, particularly in the rapidly emerging field of cancer immunotherapy.
Abstract
The genetic, functional or compositional heterogeneity of healthy and diseased tissues presents major challenges in drug discovery and development. Such heterogeneity hinders the design of accurate disease models and can confound the interpretation of biomarker levels and of patient responses to specific therapies. The complex nature of virtually all tissues has motivated the development of tools for single-cell genomic, transcriptomic and multiplex proteomic analyses. Here, we review these tools and assess their advantages and limitations. Emerging applications of single cell analysis tools in drug discovery and development, particularly in the field of oncology, are discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sakmann, B. & Neher, E. Patch clamp techniques for studying ionic channels in excitable membranes. Annu. Rev. Physiol. 46, 455–472 (1984).
Amann, R. & Fuch, B. M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 6, 339–348 (2008).
Langer-Safer, P. R., Levine, M. & Ward, D. C. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc. Natl Acad. Sci. USA 79, 4381–4385 (1982).
Herzenberg, L. A. et al. The history of the fluorescence activated cell sorter and flow cytometry: a view from Stanford. Clin. Chem. 48, 1819–1827 (2002).
Herzenberg, L. A., Julius, M. H. & Masuda, T. Demonstration that antigen-binding cells are precursors of antibody-producing cells after purification with a fluorescence-activated cell sorter. Proc. Natl Acad. Sci. USA 69, 1934–1938 (1972).
Czerkinsky, C., Nilsson, L., Nygren, H., Ouchterlony, O. & Tarkowski, A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. Immunol. Methods 65, 109–121 (1983).
Perfetto, S. P., Chattopadhyay, P. K. & Roederer, M. Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol. 4, 648–655 (2004). An illustration of the state of the art of multiplex flow cytometry.
Lu, Y. et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc. Natl Acad. Sci. USA 112, E607–E615 (2015).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Shi, Q. et al. Single cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl Acad. Sci. USA 109, 419–425 (2012). An illustration of quantitative and multiplex single-cell proteomics.
Irish, J. M. et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell 118, 217–228 (2004).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
de Bourcy, C. F. et al. A quantitative compariso of single-cell whole genome amplification methods. PLoS ONE 9, e105585 (2014).
Wang, J., Fan, H. C., Behr, B. & Quake, S. R. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150, 402–412 (2012).
Tay, S. et al. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 (2010).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1400 (2014).
Fan, H. C., Fu, G. K. & Fodor, S. P. A. Combinatorial labeling of single cells for gene expression cytometry. Science 347, 6222 (2015). Describes the CytoSeq method.
Han, L. et al. Co-detection and sequencing of genes and transcripts from the same single cells facilitated by a microfluidics platform. Sci. Rep. 4, 6485 (2014).
Mazumder, A., Tummler, K., Bathe, M. & Samson, L. D. Single-cell analysis of ribonucleotide reductase transcriptional and translational response to DNA damage. Mol. Cell. Biol. 33, 635–642 (2013).
Porichis, F. et al. High-throughput detection of miRNAs and gene-specific mRNA at the single-cell level by flow cytometry. Nat. Commun. 5, 5641 (2014).
Stahlberg, A., Thomsen, C., Ruff, D. & Aman, P. Quantitative PCR analysis of DNA, RNAs and proteins in the same single cell. Clin. Chem. 58, 1682–1691 (2012).
Xue, M. et al. Chemical methods for the simultaneous quantitation of metabolites and proteins from single cells. J. Am. Chem. Soc. 137, 4066–4069 (2015).
Wang, J. et al. Quantitating cell–cell interaction functions, with applications to glioblastoma multiforme cancer cells. Nano Lett. 12, 6101–6106 (2012).
Liadi, I. et al. Individual motile CD4+ T cells can participate in efficient multikilling through conjugation to multiple tumor cells. Cancer Immunol. Res. 3, 473–482 (2015).
Elitas, M., Brower, K., Lu, Y., Chen, J. J. & Fan, R. A microchip platform for interrogating tumormacrophage paracrine signaling at the single-cell level. Lab Chip 14, 3582–3588 (2014).
Wei, W. et al. Hypoxia induces a phase transition within a kinase signaling network in cancer cells. Proc. Natl Acad. Sci. USA 110, e1352–e1360 (2013).
Mehling, M., Frank, T., Albayrak, C. & Tay, S. Real-time tracking, retrieval and gene expression analysis of migrating human T cells. Lab Chip 15, 1276–1283 (2015).
Brouzes, E. et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl Acad. Sci. USA 106, 14195–14200 (2009).
Sendra, V. G., Lie, A., Romain, G., Agarwal, S. K. & Varadarajan, N. Detection and isolation of auto-reactive human antibodies from primary B cells. Methods 64, 153–159 (2013). Describes the microengraving technique used to identify cells with desirable proteomic signatures and then separate those cells for further analysis.
Love, J. C., Ronan, J. L., Grotenbreg, G. M., Van der Veen, A. G. & Ploegh, H. L. A microengraving method for rapid selection of single cells producing antigen specific antibodies. Nat. Biotechnol. 24, 703–707 (2006).
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Long, C. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, 6233 (2015). Reports a single-cell transcriptomics method applied to cells in a bulk environment.
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Shin, Y. S. et al. Chemistries for patterning robust DNA microbarcodes enable multiplex assays of cytoplasm proteins from single cancer cells. ChemPhysChem 11, 3063 (2010).
Amir, E. D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–548 (2013).
Shin, Y. S. et al. Protein signaling networks from single cell fluctuations and information theory profiling. Biophys. J. 100, 2378–2386 (2011).
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
Kravchenko-Balasha, N., Wang, J., Remacle, F., Levine, R. D. & Heath, J. R. Glioblastoma cellular architectures are predicted through the characterization of two-cell interactions. Proc. Natl Acad. Sci. USA 111, 6521–6526 (2014).
Bruggner, R. V., Bodenmiller, B., Dill, D. L., Tibshirani, R. J. & Nolan, G. P. Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl Acad. Sci. USA 111, E2770–E2777 (2014).
Shapiro, E., Biezuner, T. & Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 14, 618–630 (2013).
Blainey, P. C. & Quake, S. R. Dissecting genomic diversity, one cell at a time. Nat. Methods 11, 19–21 (2014).
Zhang, L. et al. Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. USA 89, 5847–5851 (1992).
Acinas, S. G., Sarma-Rupavtarm, R., Klepac-Ceraj, V. & Polz, M. F. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 71, 8966–8969 (2005).
Dean, F. B., Nelson, J. R., Giesler, T. L. & Lasken, R. S. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11, 1095–1099 (2001).
Dago, A. E. et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS ONE 9, e101777 (2014).
Zong, C., Lu, S., Chapman, A. R. & Xie, X. S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626 (2012). Reports the MALBAC technique.
Ni, X. et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl Acad. Sci. USA 110, 21083–21088 (2013).
Francis, J. M. et al. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 4, 956–971 (2014).
Dash, P. et al. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J. Clin. Invest. 121, 288–295 (2011).
Kim, S.-M. et al. Analysis of the paired TCR α- and β-chains of single human T cells. PLoS ONE 7, e37338 (2013).
Han, A., Glanville, J., Hansmann, L. & Davis, M. M. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014). Illustrates an important method that is relevant to various classes of immunotherapies.
Ng, S. B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Mamanova, L. et al. Target-enrichment strategies for next-generation sequencing. Nat. Methods 7, 111–118 (2010).
Hodges, E. et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 39, 1522–1527 (2007).
Hou, Y. et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 148, 873–885 (2012).
Xu, X. et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 148, 886–895 (2012).
Lohr, J. G. et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 32, 479–484 (2014).
Eberwine, J. et al. Analysis of gene expression in single live neurons. Proc. Natl Acad. Sci. USA 89, 3010–3014 (1992).
Tang, F. et al. mRNA-seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Islam, S. et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166 (2014).
Wu, A. R. et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods 11, 41–46 (2014).
Shalek, A. K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013).
Pollen, A. A. et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058 (2014).
Ting, D. T. et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 8, 1905–1918 (2014).
Luo, Y. et al. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell 161, 1175–1186 (2015).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). Reports on the InDrop single-cell transcriptomics method.
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Kivioja, T. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2012). A description of molecular barcoding for quantitative biology.
Grun, D., Kester, L. & van Oudenaarden, A. Validation of noise models for single-cell transcriptomics. Nat. Methods 11, 637–640 (2014). A description of the limits and possibilities of absolute quantitation from single-cell transcriptome analysis.
Polz, M. F. & Cavanaugh, C. M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64, 3724–3730 (1998).
Schloss, P. D., Gevers, D. & Westcott, S. L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6, e27310 (2011).
Suzuki, M. T. & Giovannoni, S. J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62, 625–630 (1996).
Brennecke, P. et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 10, 1093–1095 (2013).
Fu, G. K., Wilhelmy, J., Stern, D., Fan, H. C. & Fodor, S. P. A. Digital encoding of cellular mRNAs enabling precise and absolute gene expression measurement by single-molecule counting. Anal. Chem. 86, 2867–2870 (2014).
Fu, G. K. et al. Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proc. Natl Acad. Sci. USA 111, 1891–1896 (2014).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Yates, J. R., Ruse, C. I. & Nakorchevsky, A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 11, 49–79 (2009).
Picotti, P. & Aebersold, R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566 (2012).
Torres, A. J., Contento, R. L., Gordo, S., Wucherpfennig, K. W. & Love, C. L. Functional single-cell analysis of T-cell activation by supported bilayer-tethered ligands on arrays of nanowells. Lab Chip 13, 90–99 (2013).
Hughes, A. J. et al. Single-cell western blotting. Nat. Methods 11, 749–755 (2014).
Guo, M. T., Rotem, A., Heyman, J. A. & Weitz, D. A. Droplet microfluidics for high-throughput biological assays. Lab Chip 12, 2146–2155 (2012).
Huebner, A. et al. Static microdroplet arrays: a microfluidic device for droplet trapping, incubation and release for enzymatic and cell-based assays. Lab Chip 9, 692–698 (2009).
Kintses, B., van Vliet, L. D., Devenish, S. R. A. & Hollfelder, F. Microfluidic droplets: new integrated workflows for biological experiments. Curr. Opin. Chem. Biol. 14, 548–555 (2010).
Mazutis, L. et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 8, 870–891 (2013).
Yu, J. et al. Microfluidics-based single-cell functional proteomics for fundamental and applied biomedical applications. Annu. Rev. Anal. Chem. 7, 275–295 (2014). An overview of nanodroplet microfluidics methods for single-cell analysis.
McKelvey-Martin, V. J. et al. The single cell gel electrophoresis assay (comet assay): a European review. Mut. Res. 288, 47–63 (1993).
Weingeist, D. M. et al. Single-cell microarray enables high-throughput evaluation of DNA double-strand breaks and DNA repair inhibitors. Cell Cycle 12, 907–915 (2013).
Han, Q. et al. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc. Natl Acad. Sci. USA 109, 1607–1612 (2012).
Lu, Y. et al. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal. Chem. 85, 2548–2556 (2013).
Romain, G. et al. Antibody Fc-engineering improves frequency and promotes kinetic boosting of serial killing mediated by NK cells. Blood 124, 3241–3249 (2014).
Marcon, E. et al. Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat. Methods 12, 725–731 (2015). Describes the limitations of antibodies for protein detection assays.
Towbin, H., Staehelin, T. & Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA 76, 4350–4354 (1979).
Ma, C. et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat. Med. 17, 738–744 (2011).
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002). An excellent illustration of how single cells and bulk cell populations differ.
Kaern, M., Elston, T. C., Blake, W. J. & Collins, J. J. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6, 451–464 (2005).
Bengtsson, M., Stahlberg, A., Rorsman, P. & Kubista, M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 15, 1388–1392 (2005).
Ma, C. et al. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov. 3, 418–429 (2013). Illustrates the value of single-cell functional proteomics for cancer immunotherapy monitoring in patients.
Powell, A. A. et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE 7, e33788 (2012).
Halo, T. L. et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc. Natl Acad. Sci. USA 111, 17104–17109 (2014).
Dalerba, P. et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 29, 1120–1127 (2011). An example of lineage tracing via single-cell transcriptomics.
Zhao, J. L. et al. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell 14, 1–15 (2014).
Lin, L. et al. Human natural killer cells licensed by killer Ig receptor genes have an altered cytokine program that modifies CD4+ T cell function. J. Immunol. 193, 940–949 (2014).
Van Buuren, M. M., Calis, J. J. & Schumacher, T. N. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology 3, e28836 (2014).
Kellogg, R. A., Gómez-Sjöberg, R., Leyrat, A. A. & Tay, S. High-throughput microfluidic single-cell analysis pipeline for studies of signaling dynamics. Nat. Protoc. 9, 1713–1726 (2014).
Kellogg, R. A. & Tay, S. Noise facilitates transcriptional control under dynamic inputs. Cell 160, 381–392 (2015).
Yamanaka, Y. J. et al. Single-cell analysis of the dynamics and functional outcomes of interactions between human natural killer cells and target cells. Integr. Biol. 4, 1175–1184 (2012).
Gupta, P. B. et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644 (2011).
Nathanson, D. et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science 343, 72–76 (2014). A mechanism of adaptive resistance to a targeted inhibitor is revealed by single-cell analysis.
Jaiswal, S. & Weissman, I. L. Hematopoietic stem and progenitor cells and the inflammatory response. Ann. NY Acad. Sci. 1174, 118–121 (2009).
Baldridge, M. T., King, K. Y., Boles, N. C., Weksberg, D. C. & Goodell, M. A. Quiescent haematopoietic stem cells are activated by IFN-γ in response to chronic infection. Nature 465, 793–797 (2010).
Baldridge, M. T., King, K. Y. & Goodell, M. A. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 32, 57–65 (2011).
King, K. Y. & Goodell, M. A. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11, 685–692 (2011).
Nagai, Y. et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity 24, 801–812 (2006).
Bodenmiller, B. et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 30, 858–867 (2012). An illustration of CyTOF, combined with mass-label barcoding, for single-cell analysis and drug screening.
Couzin-Frankel, J. Cancer immunotherapy. Science 343, 1432 (2013).
Ledford, H. Cancer treatment: the killer within. Nature 508, 24–26 (2014).
Palucka, K. & Banchereau, J. Cancer immunotherapy via dendritic cells. Nat. Rev. Cancer 12, 265–277 (2012).
Rosenberg, S. A., Restifo, N. P., Yang, J. C., Morgan, R. A. & Dudley, M. E. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat. Rev. Cancer 8, 299–308 (2008).
Leach, D. R., Krummel, M. F. & Allison, J. P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736 (1996).
Ribas, A. & Tumeh, P. C. The future of cancer therapy: selecting patients who respond to PD-1/L1 blockade. Clin. Cancer Res. 20, 4982–4984 (2014).
Iwai, Y. et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl Acad. Sci. USA 99, 12293–12297 (2002).
Crompton, J. G., Sukumar, M. & Restifo, N. P. Uncoupling T-cell expansion from effector differentiation in cell-based immunotherapy. Immunol. Rev. 257, 264–276 (2014).
Segal, N. H. et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 68, 889–892 (2008).
Coulie, P. G., Van den Eynde, B. J., van der Bruggen, P. & Boon, T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat. Rev. Cancer 14, 135–146 (2014).
DuPage, M., Mazumdar, C., Schmidt, L. M., Cheung, A. F. & Jacks, T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 482, 405–409 (2012).
Matsushita, H. et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482, 400–404 (2012).
Robbins, P. F. et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 19, 747–752 (2013).
van Rooij, N. et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 31, e439–e442 (2013).
De Rosa, S. C., Herzenberg, L. A., Herzeberg, L. A. & Roederer, M. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat. Med. 7, 245–248 (2001).
Lee, P. P. et al. Characterization of circulating T cells specific for tumor-associated antigens in melanoma patients. Nat. Med. 5, 677–685 (1999).
Altman, J. D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
Ramachandiran, V. et al. A robust method for production of MHC tetramers with small molecule fluorophores. J. Immunol. Methods 319, 13–20 (2007).
Bakker, A. H. et al. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc. Natl Acad. Sci. USA 105, 3825–3830 (2008).
Celie, P. H. et al. UV-induced ligand exchange in MHC class I protein crystals. J. Am. Chem. Soc. 131, 12298 (2009).
Kwong, G. A. et al. Modular nucleic acid assembled p/MHC microarrays for multiplexed sorting of antigen-specific lymphophocytes. J. Am. Chem. Soc. 131, 9695–9703 (2009).
Rodenko, B. et al. Generation of peptide−MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1, 1120–1132 (2006).
Bordon, Y. Immunotherapy: checkpoint parley. Nat. Rev. Cancer 15, 3 (2015).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Rosenberg, S. A. & Restifo, N. P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015).
Gattinoni, L., Klebanoff, C. A. & Restifo, N. P. Paths to stemness: building the ultimate antitumour T cell. Nat. Rev. Cancer 12, 671–684 (2012).
Stroncek, D. F. et al. New directions in cellular therapy of cancer: a summary of the summit on cellular therapy for cancer. J. Transl. Med. 10, 48–52 (2012).
Restifo, N. P. & Gattinoni, L. Lineage relationship of effector and memory T cells. Curr. Opin. Immunol. 25, 556–563 (2013).
Polyak, K. Tumor heterogeneity confounds and illuminates: a case for Darwinian tumor evolution. Nat. Med. 20, 344–346 (2014).
Furnari, F. B., Cloughesy, T. F., Cavenee, W. K. & Mischel, P. S. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat. Rev. Cancer 15, 302–310 (2015).
Nowell, P. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Aktipis, C. A., Boddy, A. M., Gatenby, R. A., Brown, J. S. & Maley, C. C. Life history trade-offs in cancer evolution. Nat. Rev. Cancer 13, 883–892 (2013).
Almendro, V., Marusyk, A. & Polyak, K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 8, 277–302 (2013).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Semenza, G. L. Cancer–stromal cell interactions mediated by hypoxia-inducible factors promote angiogenesis, lymphangiogenesis, and metastasis. Oncogene 32, 4057–4063 (2013).
Yaffe, M. B. The scientific drunk at the lamppost: massive sequencing efforts in cancer discovery and treatment. Sci. Signal. 6, e13 (2013).
Brennan, C. W. et al. The somatic genetic landscape of glioblastoma. Cell 155, 462–477 (2013).
Chin, L. et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Inda, M.-d.-M. et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 24, 1731–1745 (2010).
Zadeh, G., Bhat, K. P. L. & Aldape, K. EGFR and EGFRvIII in glioblastoma: partners in crime. Cancer Cell 24, 403–404 (2013).
Bachoo, R. M. et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1, 269–277 (2002).
Sottoriva, A. et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl Acad. Sci. USA 110, 4009–4014 (2013).
Gill, B. J. et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc. Natl Acad. Sci. USA 111, 12550–12555 (2014).
Klages, R. et al. Nonequilibrium Statistical Physics of Small Systems: Fluctuation Relations and Beyond (Wiley, 2013).
Losick, R. & Desplan, C. Stochasticity and cell fate. Science 320, 65–68 (2008).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Aguirre-Ghiso, J. A. Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer 7, 834–846 (2007).
Kholodenko, B., Yaffe, M. B. & Kolch, W. Computational approaches for analyzing information flow in biological networks. Sci. Signal. 5, re1 (2012).
Acknowledgements
The authors acknowledge the following funding agencies and grants for support of some of the work described in this Review: The National Cancer Institute (5R01CA170689 to J.R.H. as the principal investigator (PI) and A.R. as the co-PI, and 5U54 CA119347 to J.R.H. as the PI); Stand up to Cancer Foundation (to A.R. and J.R.H.); the Cancer Research Institute (to A.R. and J.R.H.); The Ben and Catherine Ivy Foundation (to P.S.M. and J.R.H.); and the Jean Perkins Foundation (to J.R.H. as the PI).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.R.H. and A.R. are on the Scientific Advisory Board of Isoplexis, which is seeking to commercialize certain aspects of the single-cell barcode chip technology.
Glossary
- Whole-genome amplification
-
A method, first reported using PCR by Arnheim's group, for nonselectively amplifying all DNA sequences present in a given sample, including a single cell.
- Multiple displacement amplification
-
A non-PCR based, room temperature DNA amplification technique reported by Lasken's group that is commonly used for whole-genome amplification.
- Multiple annealing and looping-based amplification cycles
-
(MALBAC). A PCR-type approach reported by Xie's group in which the enzymatic amplification of cDNAs proceeds via a linear process.
- Exome sequencing
-
Genome sequencing that is limited to only the small fraction (1%) of the genome that is protein encoding.
- RNA-sequencing
-
(RNA-seq). Also called whole transcriptome shotgun sequencing, RNA-seq is a method for analysing the transcriptome of a sample using next-generation sequencing tools.
- Molecular barcoding
-
An approach through which a DNA sequence or some other molecular identifier is used as an identifier of a specific cell or a specific transcript generated by that cell.
- Cytoseq
-
A microchip-based single-cell transcriptomics method reported by Fodor's group at Cellular Research in 2015.
- inDrop
-
A nanodrop-based single-cell transcriptomics method reported by Klein and others in 2015.
- Unique molecular index
-
(UMI). A molecular barcode used to identify a specific transcript from a specific cell.
- DropSeq
-
A nanodrop-based single-cell transcriptomics method reported by Macosko and others in 2015.
- Nanodrops
-
Microfluidics methods in which individual assays are carried out in isolated nanolitre-size droplets of water, separated from one another by oil.
- Mass cytometry
-
A single cell proteomics method based on traditional flow cytometry methods but uses mass labels and mass spectrometry for protein analysis.
- Microengraving
-
A microfluidics single-cell proteomics method.
- Single-cell barcode chips
-
(SCBCs). A single-cell proteomics method
- Single-cell western blottings
-
(scWesterns). A microchip- based method for carrying out western blotting assays on single cells.
- Neoantigens
-
Small peptide fragments that contain a genetic mutation. These fragments may be recognized by T cells during an antitumour immune response.
Rights and permissions
About this article
Cite this article
Heath, J., Ribas, A. & Mischel, P. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov 15, 204–216 (2016). https://doi.org/10.1038/nrd.2015.16
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd.2015.16
This article is cited by
-
Fluorescence-based multifunctional light sheet imaging flow cytometry for high-throughput optical interrogation of live cells
Communications Physics (2024)
-
VIBRANT: spectral profiling for single-cell drug responses
Nature Methods (2024)
-
Harnessing 3D in vitro systems to model immune responses to solid tumours: a step towards improving and creating personalized immunotherapies
Nature Reviews Immunology (2024)
-
Single-Cell Proliferation Microfluidic Device for High Throughput Investigation of Replicative Potential and Drug Resistance of Cancer Cells
Cellular and Molecular Bioengineering (2023)
-
Simplifying biochemical signal transport in steady flows within a single-cell-trapping microchannel by linear low-order systems
Microfluidics and Nanofluidics (2023)