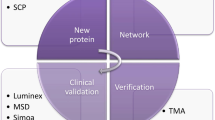
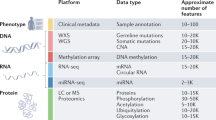
Abstract
The introduction of technologies such as mass spectrometry and protein and DNA arrays, combined with our understanding of the human genome, has enabled simultaneous examination of thousands of proteins and genes in single experiments, which has led to renewed interest in discovering novel biomarkers for cancer. The modern technologies are capable of performing parallel analyses as opposed to the serial analyses conducted with older methods, and they therefore provide opportunities to identify distinguishing patterns (signatures or portraits) for cancer diagnosis and classification as well as to predict response to therapies. Furthermore, these technologies provide the means by which new, single tumor markers could be discovered through use of reasonable hypotheses and novel analytical strategies. Despite the current optimism, a number of important limitations to the discovery of novel single tumor markers have been identified, including study design bias, and artefacts related to the collection and storage of samples. Despite the fact that new technologies and strategies often fail to identify well-established cancer biomarkers and show a bias toward the identification of high-abundance molecules, these technological advances have the capacity to revolutionize biomarker discovery. It is now necessary to focus on careful validation studies in order to identify the strategies and biomarkers that work and bring them to the clinic as early as possible.
Key Points
-
Current cancer biomarkers suffer from low diagnostic sensitivity and specificity and have not yet made a major impact in reducing cancer burden
-
The impressive growth of large-scale and high-throughput biology has resulted in increased popularity for the concept that novel biomarkers can be discovered through various emerging technologies
-
A better understanding of the mechanisms behind biomarker elevation in biological fluids may facilitate the discovery of new tumor markers
-
Some of the new promising strategies for biomarker discovery include microarray-based profiling at the DNA and mRNA level, and mass-spectrometry-based profiling at the protein or peptide level
-
Study of tumor markers that include current biomarkers or examination of fluids and tissues that are in close proximity to the tumor might also assist in identification of novel tumor markers
-
New tumor markers must undergo rigorous validation before they are introduced into routine clinical care
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jemal A et al. (2007) Cancer statistics, 2007. CA Cancer J Clin 57: 43–66
Etzioni R et al. (2003) The case for early detection. Nat Rev Cancer 3: 243–252
Hayes DF et al. (1996) Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J Natl Cancer Inst 88: 1456–1466
Jones HB (1848) On a new substance occuring in the urine with mollities ossium. Phil Trans R Soc Lond 138: 55–62
Abelev GI et al. (1963) Production of embryonal alpha-globulin by transplantable mouse hepatomas. Transplantation 1: 174–180
Gold P and Freedman SO (1965) Specific carcinoembryonic antigens of the human digestive system. J Exp Med 122: 467–481
Bast RC Jr et al. (1981) Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest 68: 1331–1337
Papsidero LD et al. (1980) A prostate antigen in sera of prostatic cancer patients. Cancer Res 40: 2428–2432
Anderson NL and Anderson NG (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 1: 845–867
Tomlins SA et al. (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648
Ono K et al. (2000) Identification by cDNA microarray of genes involved in ovarian carcinogenesis. Cancer Res 60: 5007–5011
Welsh JB et al. (2001) Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA 98: 1176–1181
Hellstrom I et al. (2003) The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma. Cancer Res 63: 3695–3700
Galgano MT et al. (2006) Comprehensive analysis of HE4 expression in normal and malignant human tissues. Mod Pathol 19: 847–853
Jarjanazi H et al. (2008) Biological implications of SNPs in signal peptide domains of human proteins. Proteins 70: 394–403
Abelev GI and Eraiser TL (1999) Cellular aspects of alpha-fetoprotein reexpression in tumors. Semin Cancer Biol 9: 95–107
Slamon DJ et al. (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235: 177–182
Shak S (1999) Overview of the trastuzumab (Herceptin) anti-HER2 monoclonal antibody clinical program in HER2-overexpressing metastatic breast cancer. Herceptin Multinational Investigator Study Group. Semin Oncol 26: 71–77
Molina R et al. (1996) C-erbB-2 oncoprotein in the sera and tissue of patients with breast cancer: utility in prognosis. Anticancer Res 16: 2295–2300
Stacker SA et al. (2002) Lymphangiogenesis and cancer metastasis. Nat Rev Cancer 2: 573–583
Quackenbush J (2006) Microarray analysis and tumor classification. N Engl J Med 354: 2463–2472
Eisen MB et al. (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868
Golub TR et al. (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286: 531–537
Perou CM et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747–752
Alizadeh AA et al. (2001) Towards a novel classification of human malignancies based on gene expression patterns. J Pathol 195: 41–52
Weigelt B et al. (2005) Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res 65: 9155–9158
Alizadeh AA et al. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511
Rosenwald A et al. (2002) The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 346: 1937–1947
Pomeroy SL et al. (2002) Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415: 436–442
Iizuka N et al. (2004) Predicting individual outcomes in hepatocellular carcinoma. Lancet 364: 1837–1839
Chen HY et al. (2007) A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 356: 11–20
van de Vijver MJ et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009
Paik S et al. (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351: 2817–2826
Pollack JR (2007) A perspective on DNA microarrays in pathology research and practice. Am J Pathol 171: 375–385
Michiels S et al. (2005) Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet 365: 488–492
Ioannidis JP (2005) Microarrays and molecular research: noise discovery. Lancet 365: 454–455
Diamandis EP . et al. (2006) National Academy of Clinical Biochemistry Guidelines: The Use of Microarrays in Cancer Diagnostics. American Association for Clinical Chemistry. 2006. Ref Type: Electronic Citation [http://www.aacc.org/NR/rdonlyres/E4CF9D42-B055-4377-A02E-F0BD3856C456/ 0/chp4a_microarray.pdf]
Domon B and Aebersold R (2006) Mass spectrometry and protein analysis. Science 312: 212–217
Wulfkuhle JD et al. (2003) Proteomic approaches to the diagnosis, treatment, and monitoring of cancer. Adv Exp Med Biol 532: 59–68
Petricoin EF et al. (2002) Use of proteomic patterns in serum to identify ovarian cancer. Lancet 359: 572–577
Li J et al. (2002) Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem 48: 1296–1304
Petricoin EF III et al. (2002) Serum proteomic patterns for detection of prostate cancer. J Natl Cancer Inst 94: 1576–1578
Chen YD et al. (2004) Artificial neural networks analysis of surface-enhanced laser desorption/ionization mass spectra of serum protein pattern distinguishes colorectal cancer from healthy population. Clin Cancer Res 10: 8380–8385
Paradis V et al. (2005) Identification of a new marker of hepatocellular carcinoma by serum protein profiling of patients with chronic liver diseases. Hepatology 41: 40–47
Tolson J et al. (2004) Serum protein profiling by SELDI mass spectrometry: detection of multiple variants of serum amyloid alpha in renal cancer patients. Lab Invest 84: 845–856
Rosty C et al. (2002) Identification of hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein I as a biomarker for pancreatic ductal adenocarcinoma by protein biochip technology. Cancer Res 62: 1868–1875
Wadsworth JT et al. (2004) Identification of patients with head and neck cancer using serum protein profiles. Arch Otolaryngol Head Neck Surg 130: 98–104
Diamandis EP (2003) Point: proteomic patterns in biological fluids: do they represent the future of cancer diagnostics? Clin Chem 49: 1272–1275
Karsan A et al. (2005) Analytical and preanalytical biases in serum proteomic pattern analysis for breast cancer diagnosis. Clin Chem 51: 1525–1528
Banks RE et al. (2005) Influences of blood sample processing on low-molecular-weight proteome identified by surface-enhanced laser desorption/ionization mass spectrometry. Clin Chem 51: 1637–1649
Ransohoff DF (2005) Lessons from controversy: ovarian cancer screening and serum proteomics. J Natl Cancer Inst 97: 315–319
Baggerly KA et al. (2005) Signal in noise: evaluating reported reproducibility of serum proteomic tests for ovarian cancer. J Natl Cancer Inst 97: 307–309
Chan DW et al. (2006) National Academy of Clinical Biochemistry Guidelines: The Use of MALDI-TOF Mass Spectrometry Profiling to Diagnose Cancer. American Association for Clinical Chemistry. 2006. Ref Type: Electronic Citation [http://www.aacc.org/NR/rdonlyres/45357D4E-FA88-4997-B8A6-74BFE31A3D49/ 0/chp4b_mass_spec.pdf]
Lopez MF et al. (2005) High-resolution serum proteomic profiling of Alzheimer disease samples reveals disease-specific, carrier-protein-bound mass signatures. Clin Chem 51: 1946–1954
Liotta LA et al. (2003) Clinical proteomics: written in blood. Nature 425: 905
Tirumalai RS et al. (2003) Characterization of the low molecular weight human serum proteome. Mol Cell Proteomics 2: 1096–1103
Harper RG et al. (2004) Low-molecular-weight human serum proteome using ultrafiltration, isoelectric focusing, and mass spectrometry. Electrophoresis 25: 1299–1306
Rai DK et al. (2004) Accurate mass measurement and tandem mass spectrometry of intact globin chains identify the low proportion variant hemoglobin Lepore-Boston-Washington from the blood of a heterozygote. J Mass Spectrom 39: 289–294
Villanueva J et al. (2006) Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest 116: 271–284
Lopez MF et al. (2007) A novel, high-throughput workflow for discovery and identification of serum carrier protein-bound peptide biomarker candidates in ovarian cancer samples. Clin Chem 53: 1067–1074
Koomen JM et al. (2005) Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J Proteome Res 4: 972–981
Diamandis EP (2006) Peptidomics for cancer diagnosis: present and future. J Proteome Res 5: 2079–2082
Borgono CA and Diamandis EP (2004) The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer 4: 876–890
Rittenhouse HG et al. (1998) Human Kallikrein 2 (hK2) and prostate-specific antigen (PSA): two closely related, but distinct, kallikreins in the prostate. Crit Rev Clin Lab Sci 35: 275–368
Diamandis EP et al. (2003) Human kallikrein 6 (hK6): a new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J Clin Oncol 21: 1035–1043
Liotta LA and Kohn EC (2001) The microenvironment of the tumour-host interface. Nature 411: 375–379
Jung YD et al. (2002) The role of the microenvironment and intercellular cross-talk in tumor angiogenesis. Semin Cancer Biol 12: 105–112
Celis JE et al. (2004) Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol Cell Proteomics 3: 327–344
Wang X et al. (2005) Autoantibody signatures in prostate cancer. N Engl J Med 353: 1224–1235
Nowell PC and Hungerford DA (1960) Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst 25: 85–109
Caprioli RM (2005) Deciphering protein molecular signatures in cancer tissues to aid in diagnosis, prognosis, and therapy. Cancer Res 65: 10642–10645
Yanagisawa K et al. (2003) Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 362: 433–439
Faca V et al. (2007) Contribution of protein fractionation to depth of analysis of the serum and plasma proteomes. J Proteome Res 6: 3558–3565
Kuick R et al. (2007) Discovery of cancer biomarkers through the use of mouse models. Cancer Lett 249: 40–48
Whiteaker JR et al. (2007) Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res 6: 3962–3975
Pepe MS et al. (2001) Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 93: 1054–1061
Bast RC Jr et al. (2001) 2000 update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J Clin Oncol 19: 1865–1878
Finne P et al. (2000) Predicting the outcome of prostate biopsy in screen-positive men by a multilayer perceptron network. Urology 56: 418–422
Stephan C et al. (2002) Multicenter evaluation of an artificial neural network to increase the prostate cancer detection rate and reduce unnecessary biopsies. Clin Chem 48: 1279–1287
Diamandis EP et al. (2002) Tumor Markers: Physiology, Pathobiology, Technology, and Clinical Applications. Washington, DC: AACC Press
Melvin KE et al. (1971) Early diagnosis of medullary carcinoma of the thyroid gland by means of calcitonin assay. N Engl J Med 285: 1115–1120
Sturgeon C (2002) Practice guidelines for tumor marker use in the clinic. Clin Chem 48: 1151–1159
Kufe D et al. (1984) Differential reactivity of a novel monoclonal antibody (DF3) with human malignant versus benign breast tumors. Hybridoma 3: 223–232
Hilkens J et al. (1984) Monoclonal antibodies against human milk-fat globule membranes detecting differentiation antigens of the mammary gland and its tumors. Int J Cancer 34: 197–206
Koprowski H et al. (1979) Colorectal carcinoma antigens detected by hybridoma antibodies. Somatic Cell Genet 5: 957–971
Ludwig JA and Weinstein JN (2005) Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer 5: 845–856
McGuire WL et al. (1977) Current status of estrogen and progesterone receptors in breast cancer. Cancer 39: 2934–2947
Coussens L et al. (1985) Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science 230: 1132–1139
Yamamoto T et al. (1986) Similarity of protein encoded by the human c-erb-B-2 gene to epidermal growth factor receptor. Nature 319: 230–234
Bagshawe KD et al. (1980) Markers in gynaecological cancer. Arch Gynecol 229: 303–310
Hill BR and Levi C (1954) Elevation of a serum component in neoplastic disease. Cancer Res 14: 513–515
Wang MC et al. (1979) Purification of a human prostate specific antigen. Invest Urol 17: 159–163
Carayanniotis G and Rao VP (1977) Searching for pathogenic epitopes in thyroglobulin: parameters and caveats. Immunol Today 18: 83–88
Acknowledgements
V Kulasingam is supported by a scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC). EP Diamandis is Associate Member of the Early Detection Research Network (EDRN) and is supported by grants from the US NIH, NSERC and the Ontario Institute for Cancer Research. The authors would like to thank Carla Borgono for her assistance in generating Figure 1. CP Vega, University of California, Irvine, CA, is the author of and is solely responsible for the content of the learning objectives, questions and answers of the Medscape-accredited continuing medical education activity associated with this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kulasingam, V., Diamandis, E. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Rev Clin Oncol 5, 588–599 (2008). https://doi.org/10.1038/ncponc1187
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncponc1187
This article is cited by
-
GBAP1 functions as a tumor promotor in hepatocellular carcinoma via the PI3K/AKT pathway
BMC Cancer (2023)
-
A systems biology approach to pathogenesis of gastric cancer: gene network modeling and pathway analysis
BMC Gastroenterology (2023)
-
Development of a predictive model to distinguish prostate cancer from benign prostatic hyperplasia by integrating serum glycoproteomics and clinical variables
Clinical Proteomics (2023)
-
Identification of potential DNA methylation biomarkers related to diagnosis in patients with bladder cancer through integrated bioinformatic analysis
BMC Urology (2023)
-
Spatial metabolomics identifies distinct tumor-specific and stroma-specific subtypes in patients with lung squamous cell carcinoma
npj Precision Oncology (2023)