Key Points
-
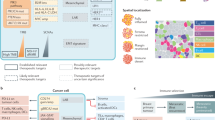
The routine diagnosis of triple-negative breast cancer (TNBC) depends on the accurate assessment of the status of the oestrogen receptor (ER), progesterone receptor (PgR) and HER2
-
Chemotherapy remains the standard therapeutic approach for TNBC at all stages, with platinum compounds having a relevant role, especially in patients harbouring BRCA1/2 mutations or 'BRCAness'
-
'Omics' technologies have provided unprecedented insights into the molecular complexity and heterogeneous clinical behaviour of TNBC but, to date, none of the newly developed molecular classifications has demonstrated clinical utility
-
Several potentially actionable molecular alterations, frequently affecting PI3K/mTOR or RAS/RAF/MEK, have been found in TNBC, but none have been confirmed as a 'driver alteration', nor have any TNBC subsets been shown to be 'addicted' to them
-
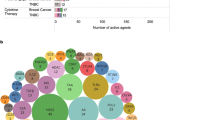
Targeted agents currently under clinical investigation in TNBC include PARP inhibitors, PI3K inhibitors, MEK inhibitors, anti-androgen therapies, heat shock protein 90 inhibitors, histone deacetylase inhibitors, and their combinations
-
TNBC is remarkably heterogeneous in terms of the tumour microenvironment; tumour lymphocyte infiltration is associated with good prognosis and a response to chemotherapy, which provides a strong rationale for testing immunotherapies in TNBC
Abstract
Chemotherapy is the primary established systemic treatment for patients with triple-negative breast cancer (TNBC) in both the early and advanced-stages of the disease. The lack of targeted therapies and the poor prognosis of patients with TNBC have fostered a major effort to discover actionable molecular targets to treat patients with these tumours. Massively parallel sequencing and other 'omics' technologies have revealed an unexpected level of heterogeneity of TNBCs and have led to the identification of potentially actionable molecular features in some TNBCs, such as germline BRCA1/2 mutations or 'BRCAness', the presence of the androgen receptor, and several rare genomic alterations. Whether these alterations are molecular 'drivers', however, has not been clearly established. A subgroup of TNBCs shows a high degree of tumour-infiltrating lymphocytes that also correlates with a lower risk of disease relapse and a higher likelihood of benefit from chemotherapy. Proof-of-principle studies with immune-checkpoint inhibitors in advanced-stage TNBC have yielded promising results, indicating the potential benefit of immunotherapy for patients with TNBC. In this Review, we discuss the most relevant molecular findings in TNBC from the past decade and the most promising therapeutic opportunities derived from these data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Banerji, S. et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405–409 (2012).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Shah, S. P. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
National Comprehensive Cancer Network. National Comprehensive Cancer Network Breast Cancer, https://www.nccn.org/store/login/login.aspx?ReturnURL=http://www.nccn.org/professionals/physician_gls/pdf/breast.pdf (2015).
Malorni, L. et al. Clinical and biologic features of triple-negative breast cancers in a large cohort of patients with long-term follow-up. Breast Cancer Res. Treat. 136, 795–804 (2012).
Hammond, M. E. et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 28, 2784–2795 (2010).
Wolff, A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 31, 3997–4013 (2013).
McCullough, A. E. et al. Central pathology laboratory review of HER2 and ER in early breast cancer: an ALTTO trial [BIG 2-06/NCCTG N063D (Alliance)] ring study. Breast Cancer Res. Treat. 143, 485–492 (2014).
Viale, G. et al. High concordance of protein (by IHC), gene (by FISH; HER2 only), and microarray readout (by TargetPrint) of ER, PgR, and HER2: results from the EORTC 10041/BIG 03–04 MINDACT trial. Ann. Oncol. 25, 816–823 (2014).
Cheang, M. C. et al. Defining breast cancer intrinsic subtypes by quantitative receptor expression. Oncology 20, 474–482 (2015).
Iwamoto, T. et al. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. J. Clin. Oncol. 30, 729–734 (2012).
Weigelt, B. & Reis-Filho, J. S. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat. Rev. Clin. Oncol. 6, 718–730 (2009).
Bertucci, F. et al. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res. 66, 4636–4644 (2006).
Huober, J. et al. Prognosis of medullary breast cancer: analysis of 13 International Breast Cancer Study Group (IBCSG) trials. Ann. Oncol. 23, 2843–2851 (2012).
Wetterskog, D. et al. Adenoid cystic carcinomas constitute a genomically distinct subgroup of triple-negative and basal-like breast cancers. J. Pathol. 226, 84–96 (2012).
Perou, C. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Sorlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009).
Hoadley, K. A. et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944 (2014).
Foulkes, W. D., Smith, I. E. & Reis-Filho, J. S. Triple-negative breast cancer. N. Engl. J. Med. 363, 1938–1948 (2010).
Prat, A. et al. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncology 18, 123–133 (2013).
Prat, A. et al. Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br. J. Cancer 111, 1532–1541 (2014).
Sikov, W. M. et al. Abstract S4-05: impact of intrinsic subtype by PAM50 and other gene signatures on pathologic complete response (pCR) rates in triple-negative breast cancer (TNBC) after neoadjuvant chemotherapy (NACT) +/- carboplatin (Cb) or bevacizumab (Bev): CALGB 40603/150709 (Allianc. Cancer Res. 75, S4-05 (2015).
Isakoff, S. J. et al. TBCRC009: a multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple-negative breast cancer. J. Clin. Oncol. 33, 1902–1909 (2015).
Tutt, A. et al. Abstract S3-01: the TNT trial: a randomized phase III trial of carboplatin (C) compared with docetaxel (D) for patients with metastatic or recurrent locally advanced triple negative or BRCA1/2 breast cancer (CRUK/07/012). Cancer Res. 75, S30-01 (2015).
Carey, L., Winer, E., Viale, G., Cameron, D. & Gianni, L. Triple-negative breast cancer: disease entity or title of convenience? Nat. Rev. Clin. Oncol. 7, 683–692 (2010).
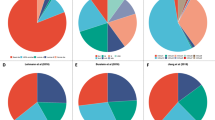
Lehmann, B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011).
Lehmann, B. D. & Pietenpol, J. A. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J. Pathol. 232, 142–150 (2014).
Masuda, H. et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. 19, 5533–5540 (2013).
Ciriello, G. et al. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 45, 1127–1133 (2013).
Lehmann, B. D. et al. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res. 16, 406 (2014).
Bose, R. et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 3, 224–237 (2013).
Jameson, J. L. & Longo, D. L. Precision medicine — personalized, problematic, and promising. N. Engl. J. Med. 372, 2229–2234 (2015).
Redig, A. J. & Jänne, P. A. Basket trials and the evolution of clinical trial design in an era of genomic medicine. J. Clin. Oncol. http://dx.doi.org/10.1200/JCO.2014.59.8433 (2015).
Stephens, P. J. et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 486, 400–404 (2012).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Wang, Y. et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014).
Venkitaraman, A. R. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu. Rev. Pathol. 4, 461–487 (2009).
Atchley, D. P. et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J. Clin. Oncol. 26, 4282–4288 (2008).
Turner, N., Tutt, A. & Ashworth, A. Hallmarks of 'BRCAness' in sporadic cancers. Nat. Rev. Cancer 4, 814–819 (2004).
Yun, M. H. & Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 (2009).
Yoshida, K. & Miki, Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 95, 866–871 (2004).
Scully, R. et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 (1997).
Taniguchi, T. et al. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100, 2414–2420 (2002).
Bhattacharyya, A., Ear, U. S., Koller, B. H., Weichselbaum, R. R. & Bishop, D. K. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275, 23899–23903 (2000).
Ding, S. L. et al. Abnormality of the DNA double-strand-break checkpoint/repair genes, ATM, BRCA1 and TP53, in breast cancer is related to tumour grade. Br. J. Cancer 90, 1995–2001 (2004).
Antoniou, A. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 72, 1117–1130 (2003).
Foulkes, W. D. et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl Cancer Inst. 95, 1482–1485 (2003).
Telli, M. L. et al. Phase II study of gemcitabine, carboplatin, and iniparib as neoadjuvant therapy for triple-negative and BRCA1/2 mutation-associated breast cancer with assessment of a tumor-based measure of genomic instability: PrECOG 0105. J. Clin. Oncol. 33, 1895–1901 (2015).
Turner, N. C. et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 26, 2126–2132 (2007).
Watkins, J. et al. Genomic complexity profiling reveals that HORMAD1 overexpression contributes to homologous recombination deficiency in triple-negative breast cancers. Cancer Discov. 5, 488–505 (2015).
Matros, E. et al. BRCA1 promoter methylation in sporadic breast tumors: relationship to gene expression profiles. Breast Cancer Res. Treat. 91, 179–186 (2005).
Turner, N. C. & Reis-Filho, J. S. Basal-like breast cancer and the BRCA1 phenotype. Oncogene 25, 5846–5853 (2006).
Couch, F. J. et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 33, 304–311 (2015).
Schnurbein, G. et al. RAD51C deletion screening identifies a recurrent gross deletion in breast cancer and ovarian cancer families. Breast Cancer Res. 15, R120 (2013).
Abkevich, V. et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 107, 1776–1782 (2012).
Birkbak, N. J. et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2, 366–375 (2012).
Popova, T. et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 72, 5454–5462 (2012).
Early Breast Cancer Trialists' Collaborative Group. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100 000 women in 123 randomised trials. Lancet 379, 432–444 (2012).
Cortazar, P. et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 384, 164–172 (2014).
Carey, L. A. et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 13, 2329–2334 (2007).
Darb-Esfahani, S. et al. Identification of biology-based breast cancer types with distinct predictive and prognostic features: role of steroid hormone and HER2 receptor expression in patients treated with neoadjuvant anthracycline/taxane-based chemotherapy. Breast Cancer Res. 11, R69 (2009).
Liedtke, C. et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 26, 1275–1281 (2008).
von Minckwitz, G. et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 30, 1796–1804 (2012).
Citron, M. L. et al. Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: first report of Intergroup Trial C9741/Cancer and Leukemia Group B Trial 9741. J. Clin. Oncol. 21, 1431–1439 (2003).
Berry, D. A. et al. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA 295, 1658–1667 (2006).
Gluz, O. et al. Triple-negative high-risk breast cancer derives particular benefit from dose intensification of adjuvant chemotherapy: results of WSG AM-01 trial. Ann. Oncol. 19, 861–870 (2008).
Henderson, I. C. et al. Improved outcomes from adding sequential paclitaxel but not from escalating doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J. Clin. Oncol. 21, 976–983 (2003).
Mazouni, C. et al. Inclusion of taxanes, particularly weekly paclitaxel, in preoperative chemotherapy improves pathologic complete response rate in estrogen receptor-positive breast cancers. Ann. Oncol. 18, 874–880 (2007).
Hayes, D. F. et al. HER2 and response to paclitaxel in node-positive breast cancer. N. Engl. J. Med. 357, 1496–1506 (2007).
Bonotto, M. et al. Measures of outcome in metastatic breast cancer: insights from a real-world scenario. Oncologist 19, 608–615 (2014).
Conlin, A. K. & Seidman, A. D. Taxanes in breast cancer: an update. Curr. Oncol. Rep. 9, 22–30 (2007).
Cardoso, F. et al. ESO-ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2). Ann. Oncol. 25, 1871–1888 (2014).
Partridge, A. H. et al. Chemotherapy and targeted therapy for women with human epidermal growth factor receptor 2-negative (or unknown) advanced breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 32, 3307–3329 (2014).
Sparano, J. A. et al. Randomized phase III trial of ixabepilone plus capecitabine versus capecitabine in patients with metastatic breast cancer previously treated with an anthracycline and a taxane. J. Clin. Oncol. 28, 3256–3263 (2010).
Kennedy, R. D., Quinn, J. E., Mullan, P. B., Johnston, P. G. & Harkin, D. P. The role of BRCA1 in the cellular response to chemotherapy. J. Natl Cancer Inst. 96, 1659–1668 (2004).
Jones, R. L. et al. The prognostic significance of Ki67 before and after neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. Treat. 116, 53–68 (2009).
Guarneri, V. et al. A prognostic model based on nodal status and Ki-67 predicts the risk of recurrence and death in breast cancer patients with residual disease after preoperative chemotherapy. Ann. Oncol. 20, 1193–1198 (2009).
Byrski, T. et al. Pathologic complete response to neoadjuvant cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res. Treat. 147, 401–405 (2014).
Silver, D. P. et al. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J. Clin. Oncol. 28, 1145–1153 (2010).
Ryan, P. D. Neoadjuvant cisplatin and bevacizumab in triple negative breast cancer (TNBC): safety and efficacy [abstract] J. Clin. Oncol. 27 (Suppl.), 551 (2009).
Sikov, W. M. et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J. Clin. Oncol. 33, 13–21 (2015).
von Minckwitz, G. et al. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol. 15, 747–756 (2014).
Symmans, W. F. et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 25, 4414–4422 (2007).
Balko, J. et al. Profiling of triple-negative breast cancers after neoadjuvant chemotherapy identifies targetable molecular alterations in the treatment-refractory residual disease. Cancer Res. 72, S3–S6 (2012).
Bauer, J. A. et al. RNA interference (RNAi) screening approach identifies agents that enhance paclitaxel activity in breast cancer cells. Breast Cancer Res. 12, R41 (2010).
Balko, J. M. et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat. Med. 18, 1052–1059 (2012).
Balko, J. M. et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 4, 232–245 (2014).
Audebert, M., Salles, B. & Calsou, P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279, 55117–55126 (2004).
Shall, S. & de Murcia, G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 460, 1–15 (2000).
Burkle, A. Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 163, 1–5 (2001).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005).
Turner, N., Tutt, A. & Ashworth, A. Targeting the DNA repair defect of BRCA tumours. Curr. Opin. Pharmacol. 5, 388–393 (2005).
Turner, N. C. et al. A synthetic lethal siRNA screen identifying genes mediating sensitivity to a PARP inhibitor. EMBO J. 27, 1368–1377 (2008).
Calabrese, C. R. et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J. Natl Cancer Inst. 96, 56–67 (2004).
Tutt, A. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376, 235–244 (2010).
Gelmon, K. A. et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 12, 852–861 (2011).
Sinha, G. Downfall of iniparib: a PARP inhibitor that doesn't inhibit PARP after all. J. Natl Cancer Inst. 106, djt447 (2014).
Rugo, H. S. et al. Veliparib/carboplatin plus standard neoadjuvant therapy for high-risk breast cancer: first efficacy results from the I-SPY 2 TRIAL [abstract]. Cancer Res. 73 (Suppl. 24), S5–02 (2013).
Dwadasi, S. et al. Cisplatin with or without rucaparib after preoperative chemotherapy in patients with triple-negative breast cancer (TNBC): Hoosier Oncology Group BRE09-146 [abstract]. J. Clin. Oncol. 32 (Suppl.), 1019 (2014).
Gucalp, A. et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic breast cancer. Clin. Cancer Res. 19, 5505–5512 (2013).
Traina, T. A. et al. Results from a phase 2 study of enzalutamide (ENZA), an androgen receptor (AR) inhibitor, in advanced AR+ triple-negative breast cancer (TNBC) [abstract]. J. Clin. Oncol. 33 (Suppl.), 1003 (2015).
Gonzalez-Angulo, A. M. et al. Androgen receptor levels and association with PIK3CA mutations and prognosis in breast cancer. Clin. Cancer Res. 15, 2472–2478 (2009).
Stemke-Hale, K. et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 68, 6084–6091 (2008).
Ibrahim, Y. H. et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2, 1036–1047 (2012).
Peng, G. et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat. Commun. 5, 3361 (2014).
Hoeflich, K. P. et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin. Cancer Res. 15, 4649–4664 (2009).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Giltnane, J. M. & Balko, J. M. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov. Med. 17, 275–283 (2014).
Cerami, E. et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Craig, D. W. et al. Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol. Cancer Ther. 12, 104–116 (2013).
Balko, J. M. et al. Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer. Cancer Res. 73, 6346–6358 (2013).
Horiuchi, D. et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J. Exp. Med. 209, 679–696 (2012).
Duncan, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012).
Infante, J. R. et al. A phase 1b study of trametinib, an oral mitogen-activated protein kinase kinase (MEK) inhibitor, in combination with gemcitabine in advanced solid tumours. Eur. J. Cancer 49, 2077–2085 (2013).
Charafe-Jauffret, E. et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 69, 1302–1313 (2009).
Creighton, C. J. et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl Acad. Sci. USA 106, 13820–13825 (2009).
Mani, S. A. et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008).
Creighton, C. J., Chang, J. C. & Rosen, J. M. Epithelial–mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J. Mammary Gland Biol. Neoplasia 15, 253–260 (2010).
Charafe-Jauffret, E. et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin. Cancer Res. 16, 45–55 (2010).
Huang, E. H. et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 69, 3382–3389 (2009).
Pontier, S. M. & Muller, W. J. Integrins in mammary-stem-cell biology and breast-cancer progression — a role in cancer stem cells? J. Cell Sci. 122, 207–214 (2009).
Britton, K. M. et al. Breast cancer, side population cells and ABCG2 expression. Cancer Lett. 323, 97–105 (2012).
Herschkowitz, J. I. et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 8, R76 (2007).
Neve, R. M. et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527 (2006).
Marotta, L. L. et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J. Clin. Invest. 121, 2723–2735 (2011).
DiMeo, T. A. et al. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial–mesenchymal transition in basal-like breast cancer. Cancer Res. 69, 5364–5373 (2009).
Bhola, N. E. et al. TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer. J. Clin. Invest. 123, 1348–1358 (2013).
Liu, S. et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 66, 6063–6071 (2006).
Harrison, H. et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 70, 709–718 (2010).
Carey, L. A. et al. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J. Clin. Oncol. 30, 2615–2623 (2012).
Baselga, J. et al. Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J. Clin. Oncol. 31, 2586–2592 (2013).
Hu, Y., Liu, J. & Huang, H. Recent agents targeting HIF-1α for cancer therapy. J. Cell. Biochem. 114, 498–509 (2013).
Sameni, M. et al. Cabozantinib (XL184) inhibits growth and invasion of preclinical TNBC models. Clin. Cancer Res. 22, 923–934 (2016).
Bianchi-Smiraglia, A., Paesante, S. & Bakin, A. V. Integrin β5 contributes to the tumorigenic potential of breast cancer cells through the Src–FAK and MEK–ERK signaling pathways. Oncogene 32, 3049–3058 (2013).
Ehrlich, P. Üeber den jetzigen Stand der Karzinomforschung. Ned. Tijdschr. Geneeskd. 5, 273–290 (1909).
Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Larkin, J. et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373, 23–34 (2015).
Rosenberg, S. A. Raising the bar: the curative potential of human cancer immunotherapy. Sci. Transl. Med. 4, 127ps8 (2012).
Schadendorf, D. et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J. Clin. Oncol. http://dx.doi.org/10.1200/JCO.2014.56.2736 (2015).
Burnet, F. M. The concept of immunological surveillance. Prog. Exp. Tumor Res. 13, 1–27 (1970).
Schreiber, R. D., Old, L. J. & Smyth, M. J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 331, 1565–1570 (2011).
Pusztai, L., Karn, T., Safonov, A., Abu-Khalaf, M. M. & Bianchini, G. New strategies in breast cancer: immunotherapy. Clin. Cancer Res. http://dx.doi.org/10.1158/1078-0432.CCR-15-1315 (2016).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Topalian, S. L., Drake, C. G. & Pardoll, D. M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015).
Brown, S. D. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 24, 743–750 (2014).
Loi, S. et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J. Clin. Oncol. 31, 860–867 (2013).
Wimberly, H. et al. PD-L1 expression correlates with tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy in breast cancer. Cancer Immunol. Res. 3, 326–332 (2015).
Ali, H. R. et al. PD-L1 protein expression in breast cancer is rare, enriched in basal-like tumours and associated with infiltrating lymphocytes. Ann. Oncol. 26, 1488–1493 (2015).
Mittendorf, E. A. et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2, 361–370 (2014).
Sabatier, R. et al. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget 6, 5449–5464 (2015).
Spranger, S. et al. Up-regulation of PD-L1, IDO, and Tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci. Transl. Med. 5, 200ra116 (2013).
Crane, C. A. et al. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene 28, 306–312 (2008).
Aaltomaa, S. et al. Lymphocyte infiltrates as a prognostic variable in female breast cancer. Eur. J. Cancer 28, 859–864 (1992).
Black, M. M., Speer, F. D. & Opler, S. R. Structural representations of tumor–host relationships in mammary carcinoma; biologic and prognostic significance. Am. J. Clin. Pathol. 26, 250–265 (1956).
Schmidt, M. et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 68, 5405–5413 (2008).
Bianchini, G. et al. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J. Clin. Oncol. 28, 4316–4323 (2010).
Desmedt, C. et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin. Cancer Res. 14, 5158–5165 (2008).
Callari, M. et al. Subtype specific metagene-based prediction of outcome after neoadjuvant and adjuvant treatment in breast cancer. Clin. Cancer Res. 22, 337–345 (2016).
Gu-Trantien, C. et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Invest. 123, 2873–2892 (2013).
Desmedt, C. et al. Multifactorial approach to predicting resistance to anthracyclines. J. Clin. Oncol. 29, 1578–1586 (2011).
West, N. R. et al. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 13, R126 (2011).
Ignatiadis, M. et al. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J. Clin. Oncol. 30, 1996–2004 (2012).
Denkert, C. et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. 33, 983–991 (2015).
Issa-Nummer, Y. et al. Prospective validation of immunological infiltrate for prediction of response to neoadjuvant chemotherapy in HER2-negative breast cancer — a substudy of the neoadjuvant GeparQuinto trial. PLoS ONE 8, e79775 (2013).
Zitvogel, L., Apetoh, L., Ghiringhelli, F. & Kroemer, G. Immunological aspects of cancer chemotherapy. Nat. Rev. Immunol. 8, 59–73 (2008).
Galluzzi, L., Senovilla, L., Zitvogel, L. & Kroemer, G. The secret ally: immunostimulation by anticancer drugs. Nat. Rev. Drug Discov. 11, 215–233 (2012).
Kang, T. H. et al. Chemotherapy acts as an adjuvant to convert the tumor microenvironment into a highly permissive state for vaccination-induced antitumor immunity. Cancer Res. 73, 2493–2504 (2013).
Dieci, M. V. et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in two phase III randomized adjuvant breast cancer trials. Ann. Oncol. 26, 1698–1704 (2015).
Loi, S. et al. RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin. Cancer Res. 22, 1499–1509 (2016).
Dieci, M. V. et al. Prognostic value of tumor-infiltrating lymphocytes on residual disease after primary chemotherapy for triple-negative breast cancer: a retrospective multicenter study. Ann. Oncol. 25, 611–618 (2014).
Teschendorff, A. E., Miremadi, A., Pinder, S. E., Ellis, I. O. & Caldas, C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 8, R157 (2007).
Rody, A. et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 11, R15 (2009).
Staaf, J. et al. Identification of subtypes in human epidermal growth factor receptor 2-positive breast cancer reveals a gene signature prognostic of outcome. J. Clin. Oncol. 28, 1813–1820 (2010).
Sabatier, R. et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res. Treat. 126, 407–420 (2011).
Rody, A. et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 13, R97 (2011).
Nagalla, S. et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 14, R34 (2013).
Loi, S. et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann. Oncol. 25, 1544–1550 (2014).
Adams, S. et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 32, 2959–2966 (2014).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann. Oncol. 26, 259–271 (2015).
Savas, P. et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat. Rev. Clin. Oncol. 13, 228–241 (2016).
Nanda, R. et al. A phase Ib study of pembrolizumab (MK-3475) in patients with advanced triple-negative breast cancer [abstract]. Cancer Res. 75, S1-09 (2015).
Emens, L. et al. Inhibition of PD-L1 by MPDL3280A leads to clinical activity in patients with metastatic triple-negative breast cancer (TNBC) [abstract]. Cancer Res. 75, 2859 (2015).
Acknowledgements
This report was supported in part by the Associazione Italiana per la Ricerca sul Cancro (AIRC) grant to G.B. (MFGA 13428) and the Fondazione Michelangelo for the advancement of the study and treatment of cancer grant to G.B.
Author information
Authors and Affiliations
Contributions
G.B., J.M.B, I.A.M. and L.G. researched data for the article. M.E.S. provided histological pictures. G.B., J.M.B, I.A.M., M.E.S. and L.G. contributed to discussing the article's content. All authors wrote, reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
G.B received honorarium from Roche. L.G. had an advisory role for Merck and Roche. J.M.B., I.A.M. and M.E.S. declare no competing interests.
Related links
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Bianchini, G., Balko, J., Mayer, I. et al. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 13, 674–690 (2016). https://doi.org/10.1038/nrclinonc.2016.66
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2016.66
This article is cited by
-
A real-world study of the effectiveness and safety of apatinib-based regimens in metastatic triple-negative breast cancer
BMC Cancer (2024)
-
Protein disulfide isomerase family member 4 promotes triple-negative breast cancer tumorigenesis and radiotherapy resistance through JNK pathway
Breast Cancer Research (2024)
-
Basal–epithelial subpopulations underlie and predict chemotherapy resistance in triple-negative breast cancer
EMBO Molecular Medicine (2024)
-
cGAS-STING pathway expression correlates with genomic instability and immune cell infiltration in breast cancer
npj Breast Cancer (2024)
-
PIWI-interacting RNA-YBX1 inhibits proliferation and metastasis by the MAPK signaling pathway via YBX1 in triple-negative breast cancer
Cell Death Discovery (2024)