Key Points
-
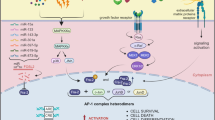
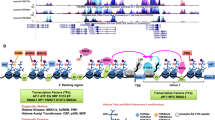
Maf proteins are bZIP transcription factors of the AP1 superfamily, like JUN and FOS.
-
During development, Maf proteins are involved early during tissue specification and later in terminal differentiation.
-
Large Maf proteins, MAFA, MAFB and MAF, are bona fide oncogenes, as demonstrated in tissue culture and animal models and in human cancer.
-
In humans, the large Maf are overexpressed in 50% of multiple myelomas and 60% of angioimmunoblastic T-cell lymphomas. In particular, MAF, MAFB and MAFA genes are translocated to the immunoglobulin heavy chain locus in 8–10% of multiple myelomas.
-
The transforming activity of large Maf proteins is context-dependent and they can occasionally display tumour suppressor-like activity in specific cellular settings.
-
Their transforming activity relies on overexpression, and is regulated by post-translational modifications, notably phosphorylation.
-
In several biological settings, large Maf induce deregulation of cell cycle, cell migration and cell–cell interactions through induction of expression of cyclin D2, ARK5 and integrin β7, respectively.
-
The oncogenic activity of Maf may result, in part, from the acquisition of novel functions.
-
A striking characteristic of Maf proteins in oncogenesis is their ability to enhance the interaction between tumour cells and stromal cells.
Abstract
Like JUN and FOS, the Maf transcription factors belong to the AP1 family. Besides their established role in human cancer — overexpression of the large Maf genes promotes the development of multiple myeloma — they can display tumour suppressor-like activity in specific cellular contexts, which is compatible with their physiological role in terminal differentiation. However, their oncogenic activity relies mostly on the acquisition of new biological functions relevant to cell transformation, the most striking characteristic of Maf oncoproteins being their ability to enhance pathological interactions between tumour cells and the stroma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Maki, Y., Bos, T. J., Davis, C., Starbuck, M. & Vogt, P. K. Avian sarcoma virus 17 carries the jun oncogene. Proc. Natl Acad. Sci. USA 84, 2848–2852 (1987).
Nishizawa, M., Kataoka, K., Goto, N., Fujiwara, K. T. & Kawai, S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc. Natl Acad. Sci. USA 86, 7711–7715 (1989). A founding paper reporting the isolation of the first member of the Maf family and its oncogenic activity in vivo .
Mariani, O. et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell 11, 361–374 (2007).
Chesi, M. et al. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood 91, 4457–4463 (1998). First demonstration of the involvement of a Maf gene in human cancer through translocation.
Kawai, S. et al. Isolation of the avian transforming retrovirus, AS42, carrying the v-maf oncogene and initial characterization of its gene product. Virology 188, 778–784 (1992).
Motohashi, H. & Yamamoto, M. Carcinogenesis and transcriptional regulation through Maf recognition elements. Cancer Sci. 98, 135–139 (2007).
Blank, V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J. Mol. Biol. 376, 913–925 (2008).
Benkhelifa, S. et al. mafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene 17, 247–254 (1998).
Ogino, H. & Yasuda, K. Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science 280, 115–118 (1998).
Kataoka, K., Fujiwara, K. T., Noda, M. & Nishizawa, M. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol. Cell. Biol. 14, 7581–7591 (1994).
Swaroop, A. et al. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc. Natl Acad. Sci. USA 89, 266–270 (1992).
Fujiwara, K. T., Kataoka, K. & Nishizawa, M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene 8, 2371–2380 (1993).
Kataoka, K. et al. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell. Biol. 15, 2180–2190 (1995).
Eferl, R. & Wagner, E. F. AP-1: a double-edged sword in tumorigenesis. Nature Rev. Cancer 3, 859–868 (2003).
Shaulian, E. & Karin, M. AP-1 as a regulator of cell life and death. Nature Cell Biol. 4, E131–136 (2002).
Vinson, C., Acharya, A. & Taparowsky, E. J. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim. Biophys. Acta 1759, 4–12 (2006).
Blank, V. & Andrews, N. C. The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci. 22, 437–441 (1997).
Dlakic, M., Grinberg, A. V., Leonard, D. A. & Kerppola, T. K. DNA sequence-dependent folding determines the divergence in binding specificities between Maf and other bZIP proteins. EMBO J. 20, 828–840 (2001).
Kerppola, T. K. & Curran, T. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene 9, 3149–3158 (1994).
Kusunoki, H. et al. Solution structure of the DNA-binding domain of MafG. Nature Struct. Biol. 9, 252–256 (2002).
Kataoka, K. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J. Biochem. 141, 775–781 (2007).
Yang, Y. & Cvekl, A. Large Maf transcription factors: cousins of AP-1 proteins and important regulators of cellular differentiation. J. Biol. Med. 23, 2–11 (2007).
Yoshida, T., Ohkumo, T., Ishibashi, S. & Yasuda, K. The 5′-AT-rich half-site of Maf recognition element: a functional target for bZIP transcription factor Maf. Nucleic Acids Res. 33, 3465–3478 (2005).
Chen, Q., Dowhan, D. H., Liang, D., Moore, D. D. & Overbeek, P. A. CREB-binding protein/p300 co-activation of crystallin gene expression. J. Biol. Chem. 277, 24081–24089 (2002).
Rocques, N. et al. GSK-3-mediated phosphorylation enhances Maf-transforming activity. Mol. Cell 28, 584–597 (2007). This work demonstrates that GSK3-mediated phosphorylation is required for Maf transforming activity and for the establishment of a gene expression programme involved in extracellular remodelling and invasion.
Friedman, J. S. et al. The minimal transactivation domain of the basic motif-leucine zipper transcription factor NRL interacts with TATA-binding protein. J. Biol. Chem. 279, 47233–47241 (2004).
Motohashi, H., O'Connor, T., Katsuoka, F., Engel, J. D. & Yamamoto, M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294, 1–12 (2002).
Kataoka, K., Noda, M. & Nishizawa, M. Transactivation activity of Maf nuclear oncoprotein is modulated by Jun, Fos and small Maf proteins. Oncogene 12, 53–62 (1996).
Motohashi, H., Katsuoka, F., Shavit, J. A., Engel, J. D. & Yamamoto, M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell 103, 865–875 (2000).
Coolen, M. et al. Phylogenomic analysis and expression patterns of large Maf genes in Xenopus tropicalis provide new insights into the functional evolution of the gene family in osteichthyans. Dev. Genes Evol. 215, 327–339 (2005).
Lecoin, L., Sii-Felice, K., Pouponnot, C., Eychene, A. & Felder-Schmittbuhl, M. P. Comparison of maf gene expression patterns during chick embryo development. Gene Expr. Patterns 4, 35–46 (2004).
Liu, Q., Ji, X., Breitman, M. L., Hitchcock, P. F. & Swaroop, A. Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene 12, 207–211 (1996).
Sevinsky, J. R., Whalen, A. M. & Ahn, N. G. Extracellular signal-regulated kinase induces the megakaryocyte GPIIb/CD41 gene through MafB/Kreisler. Mol. Cell. Biol. 24, 4534–4545 (2004).
Sakai, M. et al. Regulation of c-maf gene expression by Pax6 in cultured cells. Nucleic Acids Res. 29, 1228–1237 (2001).
Huang, K., Serria, M. S., Nakabayashi, H., Nishi, S. & Sakai, M. Molecular cloning and functional characterization of the mouse mafB gene. Gene 242, 419–426 (2000).
Raum, J. C. et al. FoxA2, Nkx2.2, and PDX-1 regulate islet β-cell-specific mafA expression through conserved sequences located between base pairs −8118 and −7750 upstream from the transcription start site. Mol. Cell. Biol. 26, 5735–5743 (2006).
Cordes, S. P. & Barsh, G. S. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79, 1025–1034 (1994).
Benkhelifa, S. et al. Phosphorylation of MafA is essential for its transcriptional and biological properties. Mol. Cell. Biol. 21, 4441–4452 (2001).
Han, S. I., Aramata, S., Yasuda, K. & Kataoka, K. MafA stability in pancreatic β cells is regulated by glucose and is dependent on its constitutive phosphorylation at multiple sites by glycogen synthase kinase 3. Mol. Cell. Biol. 27, 6593–6605 (2007).
Ochi, H., Ogino, H., Kageyama, Y. & Yasuda, K. The stability of the lens-specific Maf protein is regulated by fibroblast growth factor (FGF)/ERK signaling in lens fiber differentiation. J. Biol. Chem. 278, 537–544 (2003).
Sii-Felice, K. et al. MafA transcription factor is phosphorylated by p38 MAP kinase. FEBS Lett. 579, 3547–3554 (2005).
Bessant, D. A. et al. A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nature Genet. 21, 355–356 (1999).
Tillmanns, S. et al. SUMO modification regulates MafB-driven macrophage differentiation by enabling Myb-dependent transcriptional repression. Mol. Cell. Biol. 27, 5554–5564 (2007).
Kataoka, K., Nishizawa, M. & Kawai, S. Structure-function analysis of the maf oncogene product, a member of the b-Zip protein family. J. Virol. 67, 2133–2141 (1993).
Nishizawa, M., Kataoka, K. & Vogt, P. K. MafA has strong cell transforming ability but is a weak transactivator. Oncogene 22, 7882–7890 (2003).
Pouponnot, C. et al. Cell context reveals a dual role for Maf in oncogenesis. Oncogene 25, 1299–1310 (2006). This study shows that Maf transforming activity is context-dependent and in some settings Maf proteins can also display anti-oncogenic activity.
Morito, N. et al. Overexpression of c-Maf contributes to T-cell lymphoma in both mice and human. Cancer Res. 66, 812–819 (2006). This study reports the unique demonstration of MAF oncogenic activity in a transgenic mouse model and MAF implication in AITL in humans.
Murakami, Y. I. et al. c-Maf expression in angioimmunoblastic T-cell lymphoma. Am. J. Surg. Pathol. 31, 1695–1702 (2007).
Hurt, E. M. et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 5, 191–199 (2004). A landmark paper showing that MAF is causative of MM pathology and increases the interaction between tumour cells and stroma.
Kataoka, K., Shioda, S., Yoshitomo-Nakagawa, K., Handa, H. & Nishizawa, M. Maf and Jun nuclear oncoproteins share downstream target genes for inducing cell transformation. J. Biol. Chem. 276, 36849–36856 (2001).
Mattioli, M. et al. Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene 24, 2461–2473 (2005).
Suzuki, A. et al. ARK5 is transcriptionally regulated by the Large-MAF family and mediates IGF-1-induced cell invasion in multiple myeloma: ARK5 as a new molecular determinant of malignant multiple myeloma. Oncogene 24, 6936–6944 (2005).
Zhan, F. et al. The molecular classification of multiple myeloma. Blood 108, 2020–2028 (2006). Identification of a Maf signature by gene expression profiling on a large cohort of patients with MM.
Monteiro, P. et al. AhR- and c-maf-dependent induction of β7-integrin expression in human macrophages in response to environmental polycyclic aromatic hydrocarbons. Biochem. Biophys. Res. Commun. 358, 442–448 (2007).
Kuehl, W. M. & Bergsagel, P. L. Multiple myeloma: evolving genetic events and host interactions. Nature Rev. Cancer 2, 175–187 (2002). A comprehensive review of MM pathogenesis.
Chng, W. J., Glebov, O., Bergsagel, P. L. & Kuehl, W. M. Genetic events in the pathogenesis of multiple myeloma. Best Pract. Res. Clin. Haematol. 20, 571–596 (2007).
Hideshima, T., Mitsiades, C., Tonon, G., Richardson, P. G. & Anderson, K. C. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nature Rev. Cancer 7, 585–598 (2007).
Kuehl, W. M. & Bergsagel, P. L. Early genetic events provide the basis for a clinical classification of multiple myeloma. Hematol. Am. Soc. Hematol. Educ. Program 346–352 (2005).
Boersma-Vreugdenhil, G. R. et al. The recurrent translocation t(14;20)(q32;q12) in multiple myeloma results in aberrant expression of MAFB: a molecular and genetic analysis of the chromosomal breakpoint. Br. J. Haematol. 126, 355–363 (2004).
Hanamura, I. et al. Ectopic expression of MAFB gene in human myeloma cells carrying (14;20)(q32;q11) chromosomal translocations. Jpn. J. Cancer Res. 92, 638–644 (2001).
Hanamura, I. et al. Identification of three novel chromosomal translocation partners involving the immunoglobulin loci in newly diagnosed myeloma and human myeloma cell lines. Blood 106, abstract 1552 (2005).
Avet-Loiseau, H. et al. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood 99, 2185–2191 (2002).
Zhan, F. et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood 109, 1692–1700 (2007).
Bergsagel, P. L. et al. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 106, 296–303 (2005).
Moreaux, J. et al. TACI expression is associated with a mature bone marrow plasma cell signature and C-MAF overexpression in human myeloma cell lines. Haematologica 92, 803–811 (2007).
Carrasco, D. R. et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 11, 349–360 (2007).
Shaughnessy, J. D. Jr et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109, 2276–2284 (2007).
Yaccoby, S. et al. Antibody-based inhibition of DKK1 suppresses tumor-induced bone resorption and multiple myeloma growth in vivo. Blood 109, 2106–2111 (2007).
Robbiani, D. F. et al. Osteopontin dysregulation and lytic bone lesions in multiple myeloma. Hematol. Oncol. 25, 16–20 (2007).
Reza, H. M., Nishi, H., Kataoka, K., Takahashi, Y. & Yasuda, K. L-Maf regulates p27kip1 expression during chick lens fiber differentiation. Differentiation 75, 737–744 (2007).
Ring, B. Z., Cordes, S. P., Overbeek, P. A. & Barsh, G. S. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development 127, 307–317 (2000).
Yoshida, K. et al. Proliferation in the posterior region of the lens of c-maf−/− mice. Curr. Eye Res. 23, 116–119 (2001).
Suzuki, A. et al. ARK5 is a tumor invasion-associated factor downstream of Akt signaling. Mol. Cell. Biol. 24, 3526–3535 (2004).
Kienast, J. & Berdel, W. E. c-maf in multiple myeloma: an oncogene enhancing tumor-stroma interactions. Cancer Cell 5, 109–110 (2004).
Li, M. A., Alls, J. D., Avancini, R. M., Koo, K. & Godt, D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nature Cell Biol. 5, 994–1000 (2003).
Zhang, C. et al. MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 25, 4969–4976 (2005).
Karnoub, A. E. et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449, 557–563 (2007).
Cohen, P. & Goedert, M. GSK3 inhibitors: development and therapeutic potential. Nature Rev. Drug Discov. 3, 479–487 (2004).
Mao, X. et al. A chemical biology screen identifies glucocorticoids that regulate c-maf expression by increasing its proteasomal degradation through up-regulation of ubiquitin. Blood 110, 4047–4054 (2007).
Yoh, K. et al. Transgenic over-expression of MafK suppresses T cell proliferation and function in vivo. Genes Cells 6, 1055–1066 (2001).
Amit, I. et al. A module of negative feedback regulators defines growth factor signaling. Nature Genet. 39, 503–512 (2007).
Dhakshinamoorthy, S. & Jaiswal, A. K. c-Maf negatively regulates ARE-mediated detoxifying enzyme genes expression and anti-oxidant induction. Oncogene 21, 5301–5312 (2002).
Kataoka, K., Yoshitomo-Nakagawa, K., Shioda, S. & Nishizawa, M. A set of Hox proteins interact with the Maf oncoprotein to inhibit its DNA binding, transactivation, and transforming activities. J. Biol. Chem. 276, 819–826 (2001).
van Dam, H. et al. Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev. 12, 1227–1239 (1998).
Hegde, S. P., Zhao, J., Ashmun, R. A. & Shapiro, L. H. c-Maf induces monocytic differentiation and apoptosis in bipotent myeloid progenitors. Blood 94, 1578–1589 (1999).
Peng, S., Lalani, S., Leavenworth, J. W., Ho, I. C. & Pauza, M. E. c-Maf interacts with c-Myb to down-regulate Bcl-2 expression and increase apoptosis in peripheral CD4 cells. Eur. J. Immunol. 37, 2868–2880 (2007).
Hale, T. K. et al. Maf transcriptionally activates the mouse p53 promoter and causes a p53-dependent cell death. J. Biol. Chem. 275, 17991–17999 (2000).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Kim, J. I., Ho, I. C., Grusby, M. J. & Glimcher, L. H. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity 10, 745–751 (1999).
Mueller, M. M. & Fusenig, N. E. Friends or foes — bipolar effects of the tumour stroma in cancer. Nature Rev. Cancer 4, 839–849 (2004).
Kerppola, T. K. & Curran, T. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene 9, 675–684 (1994).
Matsushima-Hibiya, Y., Nishi, S. & Sakai, M. Rat maf-related factors: the specificities of DNA binding and heterodimer formation. Biochem. Biophys. Res. Commun. 245, 412–418 (1998).
Newman, J. R. & Keating, A. E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300, 2097–2101 (2003).
Mechta-Grigoriou, F., Giudicelli, F., Pujades, C., Charnay, P. & Yaniv, M. c-jun regulation and function in the developing hindbrain. Dev. Biol. 258, 419–431 (2003).
Blanchi, B. et al. MafB deficiency causes defective respiratory rhythmogenesis and fatal central apnea at birth. Nature Neurosci. 6, 1091–1100 (2003).
Aziz, A. et al. Development of macrophages with altered actin organization in the absence of MafB. Mol. Cell. Biol. 26, 6808–6818 (2006).
Moriguchi, T. et al. MafB is essential for renal development and F4/80 expression in macrophages. Mol. Cell. Biol. 26, 5715–5727 (2006).
Sadl, V. et al. The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev. Biol. 249, 16–29 (2002).
Artner, I. et al. MafB is required for islet β cell maturation. Proc. Natl Acad. Sci. USA 104, 3853–3858 (2007).
Nishimura, W. et al. Preferential reduction of beta cells derived from Pax6–MafB pathway in MafB deficient mice. Dev. Biol. 314, 443–456 (2008).
Kawauchi, S. et al. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem. 274, 19254–19260 (1999).
Kim, J. I., Li, T., Ho, I. C., Grusby, M. J. & Glimcher, L. H. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc. Natl Acad. Sci. USA 96, 3781–3785 (1999).
MacLean, H. E. et al. Absence of transcription factor c-maf causes abnormal terminal differentiation of hypertrophic chondrocytes during endochondral bone development. Dev. Biol. 262, 51–63 (2003).
Mears, A. J. et al. Nrl is required for rod photoreceptor development. Nature Genet. 29, 447–452 (2001).
Kataoka, K., Noda, M. & Nishizawa, M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 14, 700–712 (1994).
Acknowledgements
The authors would like to thank the members of the laboratory for helpful discussions and C. Tran Quang, M. Mhlanga and G. Kirchweger for critical reading of the manuscript. A.E.'s laboratory is funded by grants from the INCa (French National Institute of Cancer), the Association pour la Recherche sur le Cancer and the Ligue Nationale contre le Cancer.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
European Bioinformatics Institute
National Cancer Institute
National Cancer Institute Drug Dictionary
Pfam
OMIM
FURTHER INFORMATION
Angioimmunoblastic T-cell lymphoma
National Laboratory for Computational Science and Engineering
Glossary
- Immediate early genes
-
Genes that show rapid and transient expression in the absence of de novo protein synthesis following stimulation by extracellular signals such as growth factors.
- Pulverulent cataract
-
Cataract is a cause of blindness resulting from an opacity that develops in the crystalline lens of the eye or in its envelope. Pulverulent cataracts are characterized by powdery (pulverised) opacities that may be found in any part of the lens.
- Clumped pigmentary retinal degeneration
-
An inherited retinal disorder also known as the enhanced S-cone syndrome or Goldmann–Favre syndrome, which involves retinal degeneration accompanied by clusters of large, clumped pigment deposits in the peripheral fundus at the level of the retinal pigment epithelium.
- Retinitis pigmentosa
-
Refers to a group of inherited diseases causing retinal degeneration characterized by abnormalities of the photoreceptors (rods and cones) or of the retinal pigment epithelium, leading to progressive vision loss.
- Angioimmunoblastic T-cell lymphoma
-
An aggressive peripheral T-cell lymphoma previously known as angioimmunoblastic lymphadenopathy with dysproteinaemia. It represents disease ranging from a hyperplastic but still benign immune reaction to frank malignant lymphoma.
- Extracellular matrix
-
(ECM). Complex three-dimensional network of macromolecular protein fibres as well as non-fibrous proteoglycans that is present between clusters of cells in the stroma of all tissues. The ECM provides architectural structure and strength and contextual information for cellular communication, adhesion and migration.
Rights and permissions
About this article
Cite this article
Eychène, A., Rocques, N. & Pouponnot, C. A new MAFia in cancer. Nat Rev Cancer 8, 683–693 (2008). https://doi.org/10.1038/nrc2460
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc2460
This article is cited by
-
Macrophage re-programming by JAK inhibitors relies on MAFB
Cellular and Molecular Life Sciences (2024)
-
The role and regulation of Maf proteins in cancer
Biomarker Research (2023)
-
Integrative analysis reveals early epigenetic alterations in high-grade serous ovarian carcinomas
Experimental & Molecular Medicine (2023)
-
Macrophage subsets and their role: co-relation with colony-stimulating factor-1 receptor and clinical relevance
Immunologic Research (2023)
-
MAF amplification licenses ERα through epigenetic remodelling to drive breast cancer metastasis
Nature Cell Biology (2023)