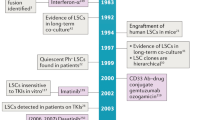
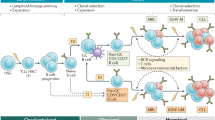
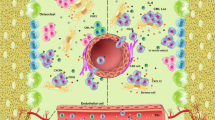
Key Points
-
Chronic myeloid leukaemia (CML) can be considered, for some aspects, a model for malignant tumours that evolve through a multi-step pathogenetic process. In its natural history, CML is usually diagnosed in chronic phase (CP) and then progresses through an accelerated phase to a nearly invariably fatal blast crisis (BC).
-
The mechanisms of transformation to BC are varied and not entirely understood. So far the best characterized include differentiation arrest, genomic instability, telomere shortening and loss of tumour-suppressor functions.
-
The differentiation arrest of the transformed BC clone is caused by the suppression of translation of the transcription factor CEBPα induced by the BCR-ABL oncoprotein in CML cells, through increased stability of the heterogenous nuclear ribonucleoprotein E2 translational regulator.
-
The increased genomic instability in CML cells is the result of their reduced capacity to survey the genome for DNA damage and to correctly repair DNA lesions, which leads to the accumulation of deleterious mutations in genes that are essential for maintaining normal cell physiology and maturation.
-
The impairment of several tumour-suppressor genes has been variably associated with BC of CML, including TP53, retinoblastoma 1, CDKN2A and others. Protein phosphatase 2a (PP2A) has an important role, as it is inhibited by BCR-ABL in BC cells by the post-transcriptional upregulation of SET, a phosphoprotein that is frequently overexpressed in other leukaemias and solid tumours.
-
Recent expression profiling of cells from different stages of CML has uncovered several genes that are differentially expressed in BC and in extremely aggressive forms of the disease. These are likely to be associated with the kinetics of blastic transformation.
-
It is envisaged that the identification of the most pathologically relevant genetic lesions for the development of BC will allow the early diagnosis of impending disease progression and the design of new treatment strategies to halt this process. Such an approach can become the paradigm for therapy decision making in other types of cancer.
Abstract
Chronic myeloid leukaemia (CML) can be considered as a paradigm for neoplasias that evolve through a multi-step process. CML is also one of the best examples of a disease that can be targeted by molecular therapy; however, the success of new 'designer drugs' is largely restricted to the chronic phase of the disease. If not cured at this stage, CML invariably progresses and transforms into an acute-type leukaemia undergoing a 'blast crisis'. The causes of this transformation are still poorly understood. What mechanisms underlie this progression, and are they shared by other common cancers?
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nowell, P. C. & Hungerford, D. A. A minute chromosome in human chronic granulocytic leukemia. Science 132, 1497 (1960).
Rowley, J. D. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 (1973).
Lugo, T. G., Pendergast, A. M., Muller, A. J. & Witte, O. N. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science 247, 1079–1082 (1990).
McLaughlin, J., Chianese, E. & Witte, O. N. In vitro transformation of immature hematopoietic cells by the P210 BCR/ABL oncogene product of the Philadelphia chromosome. Proc. Natl Acad. Sci. USA 84, 6558–6562 (1987).
Daley, G. Q., Van Etten, R. A. & Baltimore, D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science 247, 824–830 (1990).
Druker, B. J. et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nature Med. 2, 561–566 (1996).
Druker, B. J. et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 344, 1031–1037 (2001).
Bartram, C. R. et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature 306, 277–280 (1983).
Groffen, J. et al. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell 36, 93–99 (1984).
Shtivelman, E., Lifshitz, B., Gale, R. P., Roe, B. A. & Canaani, E. Alternative splicing of RNAs transcribed from the human abl gene and from the BCR-ABL fused gene. Cell 47, 277–284 (1986).
Deininger, M. W., Goldman, J. M. & Melo, J. V. The molecular biology of chronic myeloid leukemia. Blood 96, 3343–3356 (2000).
Wetzler, M. et al. Subcellular localization of Bcr, Abl, and Bcr-Abl proteins in normal and leukemic cells and correlation of expression with myeloid differentiation. J. Clin. Invest. 92, 1925–1939 (1993).
Van Etten, R. A., Jackson, P. & Baltimore, D. The mouse type IV c-abl gene product is a nuclear protein, and activation of transforming ability is associated with cytoplasmic localization. Cell 58, 669–678 (1989).
Calabretta, B. & Perrotti, D. The biology of CML blast crisis. Blood 103, 4010–4022 (2004).
Dumon, S. et al. IL-3 dependent regulation of Bcl-xL gene expression by STAT5 in a bone marrow derived cell line. Oncogene 18, 4191–4199 (1999).
Horita, M. et al. Blockade of the bcr-Abl kinase activity induces apoptosis of chronic myelogenous leukemia cells by suppressing signal transducer and activator of transcription 5-dependent expression of bcl-x(L). J. Exp. Med. 191, 977–984 (2000).
Matsumura, I. et al. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 18, 1367–1377 (1999).
de Groot, R. P., Raaijmakers, J. A., Lammers, J. W. & Koenderman, L. STAT5-dependent cyclinD1 and Bcl-xL expression in Bcr-Abl-transformed cells. Mol. Cell Biol. Res. Commun. 3, 299–305 (2000).
Klucher, K. M., Lopez, D. V. & Daley, G. Q. Secondary mutation maintains the transformed state in BaF3 cells with inducible BCR/ABL expression. Blood 91, 3927–3934 (1998).
Cambier, N., Chopra, R., Strasser, A., Metcalf, D. & Elefanty, A. G. BCR-ABL activates pathways mediating cytokine independence and protection against apoptosis in murine hematopoietic cells in a dose-dependent manner. Oncogene 16, 335–348 (1998).
Issaad, C. et al. Biological effects induced by variable levels of BCR-ABL protein in the pluripotent hematopoietic cell line UT-7. Leukemia 14, 662–670 (2000).
Barnes, D. J., Schultheis, B., Adedeji, S. & Melo, J. V. Dose-dependent effects of Bcr-Abl in cell line models of different stages of chronic myeloid leukemia. Oncogene 24, 6432–6440 (2005). In this paper the gene dosage effects of BCR-ABL in cell lines are shown to model the phenotypic characteristics of chronic and advanced phase CML.
Barnes, D. J. et al. Bcr-Abl expression levels determine the rate of development of resistance to imatinib mesylate in chronic myeloid leukemia. Cancer Res. 65, 8912–8919 (2005).
Gaiger, A. et al. Increase of bcr-abl chimeric mRNA expression in tumor cells of patients with chronic myeloid leukemia precedes disease progression. Blood 86, 2371–2378 (1995).
Guo, J. Q., Wang, J. Y. & Arlinghaus, R. B. Detection of BCR-ABL proteins in blood cells of benign phase chronic myelogenous leukemia patients. Cancer Res. 51, 3048–3051 (1991).
Elmaagacli, A. H., Beelen, D. W., Opalka, B., Seeber, S. & Schaefer, U. W. The amount of BCR-ABL fusion transcripts detected by the real-time quantitative polymerase chain reaction method in patients with Philadelphia chromosome positive chronic myeloid leukemia correlates with the disease stage. Ann. Hematol. 79, 424–431 (2000).
Lin, F., van Rhee, F., Goldman, J. M. & Cross, N. C. Kinetics of increasing BCR-ABL transcript numbers in chronic myeloid leukemia patients who relapse after bone marrow transplantation. Blood 87, 4473–4478 (1996).
Sell, S. Stem cell origin of cancer and differentiation therapy. Crit. Rev. Oncol. Hematol. 51, 1–28 (2004).
Perrotti, D. et al. BCR-ABL suppresses C/EBPα expression through inhibitory action of hnRNP E2. Nature Genet. 30, 48–58 (2002). The authors show how BCR-ABL suppresses the translation of CEBPα through HNRNPE2, and thereby reveal a mechanism that contributes to differentiation arrest in CML.
Smith, L. T., Hohaus, S., Gonzalez, D. A., Dziennis, S. E. & Tenen, D. G. PU. 1 (Spi-1) and C/EBPα regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood 88, 1234–1247 (1996).
Wagner, K. et al. Absence of the transcription factor CCAAT enhancer binding protein alpha results in loss of myeloid identity in bcr/abl-induced malignancy. Proc. Natl Acad. Sci. USA 103, 6338–6343 (2006).
Pabst, T. et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPα), in acute myeloid leukemia. Nature Genet. 27, 263–270 (2001).
Pabst, T. et al. Mutations of the myeloid transcription factor CEBPA are not associated with the blast crisis of chronic myeloid leukaemia. Br. J. Haematol. 133, 400–402 (2006).
Ferrari-Amorotti, G. et al. Leukemogenesis induced by wild-type and STI571-resistant BCR/ABL is potently suppressed by C/EBPα. Blood 108, 1353–1362 (2006).
Guerzoni, C. et al. Inducible activation of CEBPB, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood 107, 4080–4089 (2006).
Zhang, D. E. et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein a-deficient mice. Proc. Natl Acad. Sci. USA 94, 569–574 (1997).
Cuenco, G. M. & Ren, R. Cooperation of BCR-ABL and AML1/MDS1/EVI1 in blocking myeloid differentiation and rapid induction of an acute myelogenous leukemia. Oncogene 20, 8236–8248 (2001).
Dash, A. B. et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc. Natl Acad. Sci. USA 99, 7622–7627 (2002).
Nucifora, G. et al. Involvement of the AML1 gene in the t(3;21) in therapy-related leukemia and in chronic myeloid leukemia in blast crisis. Blood 81, 2728–2734 (1993).
Mitani, K. et al. Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 13, 504–510 (1994).
Ahuja, H. G., Popplewell, L., Tcheurekdjian, L. & Slovak, M. L. NUP98 gene rearrangements and the clonal evolution of chronic myelogenous leukemia. Genes Chromosomes Cancer 30, 410–415 (2001).
Yamamoto, K., Nakamura, Y., Saito, K. & Furusawa, S. Expression of the NUP98/HOXA9 fusion transcript in the blast crisis of Philadelphia chromosome-positive chronic myelogenous leukaemia with t(7;11)(p15;p15). Br. J. Haematol. 109, 423–426 (2000).
Jamieson, C. H. et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351, 657–667 (2004). A significant work showing that the pool of granulocyte-macrophage progenitors have properties, such as a high capacity for self-renewal, that make them candidates for being leukaemic stem cells.
Reya, T. et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414 (2003).
Willert, K. et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 (2003).
Notari, M. et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood 107, 2507–2516 (2006).
Huntly, B. J. & Gilliland, D. G. Blasts from the past: new lessons in stem cell biology from chronic myelogenous leukemia. Cancer Cell 6, 199–201 (2004).
Korinek, V. et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 275, 1784–1787 (1997).
Rask, K. et al. Wnt-signalling pathway in ovarian epithelial tumours: increased expression of β-catenin and GSK3β. Br. J. Cancer 89, 1298–1304 (2003).
Uematsu, K. et al. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene 22, 7218–7221 (2003).
Johansson, B., Fioretos, T. & Mitelman, F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 107, 76–94 (2002).
Melo, J. V., Kumberova, A., van Dijk, A. G., Goldman, J. M. & Yuille, M. R. Investigation on the role of the ATM gene in chronic myeloid leukaemia. Leukemia 15, 1448–1450 (2001).
Dierov, J., Dierova, R. & Carroll, M. BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S-phase checkpoint. Cancer Cell 5, 275–285 (2004).
Vigneri, P. & Wang, J. Y. Induction of apoptosis in chronic myelogenous leukemia cells through nuclear entrapment of BCR-ABL tyrosine kinase. Nature Med. 7, 228–234 (2001).
Bartek, J. & Lukas, J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3, 421–429 (2003).
Costanzo, V. et al. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11, 203–213 (2003).
Dierov, J. K., Schoppy, D. W. & Carroll, M. CML progenitor cells have chromsomal instability and display increased DNA damage at DNA fragile sites. Blood 106, 563A (2005).
Miki, Y. et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 (1994).
Risch, H. A. et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J. Natl Cancer Inst. 98, 1694–1706 (2006).
Wang, Y. et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14, 927–939 (2000). An outstanding study in which many genome surveillance proteins are shown to associate with BRCA1 to form a 'super complex'.
Deutsch, E. et al. Down-regulation of BRCA1 in BCR-ABL-expressing hematopoietic cells. Blood 101, 4583–4588 (2003).
Yarden, R. I., Pardo-Reoyo, S., Sgagias, M., Cowan, K. H. & Brody, L. C. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nature Genet. 30, 285–289 (2002).
Nieborowska-Skorska, M. et al. ATR-Chk1 axis protects BCR/ABL leukemia cells from the lethal effect of DNA double-strand breaks. Cell Cycle 5, 994–1000 (2006).
Khanna, K. K. & Jackson, S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nature Genet. 27, 247–254 (2001). An essential review of DNA double-strand breaks and how errors resulting from defective repair lead to cancer.
Deutsch, E. et al. BCR-ABL down-regulates the DNA repair protein DNA-PKcs. Blood 97, 2084–2090 (2001).
Gaymes, T. J., Mufti, G. J. & Rassool, F. V. Myeloid leukemias have increased activity of the nonhomologous end- joining pathway and concomitant DNA misrepair that is dependent on the Ku70/86 heterodimer. Cancer Res. 62, 2791–2797 (2002).
Brady, N., Gaymes, T. J., Cheung, M., Mufti, G. J. & Rassool, F. V. Increased error-prone NHEJ activity in myeloid leukemias is associated with DNA damage at sites that recruit key nonhomologous end-joining proteins. Cancer Res. 63, 1798–1805 (2003).
Bentley, J., Diggle, C. P., Harnden, P., Knowles, M. A. & Kiltie, A. E. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 32, 5249–5259 (2004).
Fu, Y. P. et al. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res. 63, 2440–2446 (2003).
Slupianek, A. et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell 8, 795–806 (2001).
Amarante, M. G. et al. Bcr-Abl exerts its antiapoptotic effect against diverse apoptotic stimuli through blockage of mitochondrial release of cytochrome C and activation of caspase-3. Blood 91, 1700–1705 (1998).
Dubrez, L. et al. BCR-ABL delays apoptosis upstream of procaspase-3 activation. Blood 91, 2415–2422 (1998).
Nowicki, M. O. et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood 104, 3746–3753 (2004). An elegant study showing that BCR-ABL both induces DNA-damaging reactive oxygen species and stimulates unfaithful DNA repair.
Slupianek, A., Nowicki, M. O., Koptyra, M. & Skorski, T. BCR/ABL modifies the kinetics and fidelity of DNA double-strand breaks repair in hematopoietic cells. DNA Repair (Amst.) 5, 243–250 (2006).
Koptyra, M. & Skorski, T. Fanconi anemia D2 (FANCD2) is required to overcome reactive oxygen species (ROS)-induced DNA double-strand breaks in BCR/ABL-positive leukemias. Blood 108, 601a (2006).
Rink, L. et al. Enhanced phosphorylation of Nbs1, a member of DNA repair/checkpoint complex RAD50-Mre11-Nbs1, can be targeted simultaneously with BCR/ABL kinase to eliminate leukemia cells. Blood 108, 603a (2006).
Slupianek, A., Jozwiakowski, S., Gurdek, E., Nowicki, M. O. & Skorski, T. BCR/ABL regulates the expression and interacts with Werner syndrome helicase/exonuclease to modulate its biochemical properties. Blood 106, 805A (2005).
Erkko, H. et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature 446, 316–319 (2007).
de Laat, W. L., Jaspers, N. G. & Hoeijmakers, J. H. Molecular mechanism of nucleotide excision repair. Genes Dev. 13, 768–785 (1999). A comprehensive review of one of the main DNA repair mechanisms.
Canitrot, Y. et al. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood 102, 2632–2637 (2003).
Takeda, N., Shibuya, M. & Maru, Y. The BCR-ABL oncoprotein potentially interacts with the xeroderma pigmentosum group B protein. Proc. Natl Acad. Sci. USA 96, 203–207 (1999).
Aboussekhra, A. et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80, 859–868 (1995).
Matakidou, A., Eisen, T., Fleischmann, C., Bridle, H. & Houlston, R. S. Evaluation of xeroderma pigmentosum XPA, XPC, XPD, XPF, XPB, XPG and DDB2 genes in familial early-onset lung cancer predisposition. Int. J. Cancer 119, 964–967 (2006).
Loeb, L. A. A mutator phenotype in cancer. Cancer Res. 61, 3230–3239 (2001).
Loeb, L. A., Loeb, K. R. & Anderson, J. P. Multiple mutations and cancer. Proc. Natl Acad. Sci. USA 100, 776–781 (2003).
Salloukh, H. F. & Laneuville, P. Increase in mutant frequencies in mice expressing the BCR-ABL activated tyrosine kinase. Leukemia 14, 1401–1404 (2000).
Brain, J. M., Goodyer, N. & Laneuville, P. Measurement of genomic instability in preleukemic P190BCR/ABL transgenic mice using inter-simple sequence repeat polymerase chain reaction. Cancer Res. 63, 4895–4898 (2003).
Canitrot, Y. et al. Mutator phenotype of BCR–ABL transfected Ba/F3 cell lines and its association with enhanced expression of DNA polymerase β. Oncogene 18, 2676–2680 (1999).
Koptyra, M. et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood 108, 319–327 (2006). The first study to provide evidence for a mechanism by which BCR-ABL can induce self-mutagenesis.
Canitrot, Y. et al. Enhanced expression and activity of DNA polymerase β in chronic myelogenous leukemia. Anticancer Res. 26, 523–525 (2006).
Hoeijmakers, J. H. Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 (2001).
Hastie, N. D. et al. Telomere reduction in human colorectal carcinoma and with ageing. Nature 346, 866–868 (1990).
Ohyashiki, J. H. et al. Telomere shortening associated with disease evolution patterns in myelodysplastic syndromes. Cancer Res. 54, 3557–3560 (1994).
Artandi, S. E. & DePinho, R. A. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10, 39–46 (2000).
Drummond, M., Lennard, A., Brummendorf, T. & Holyoake, T. Telomere shortening correlates with prognostic score at diagnosis and proceeds rapidly during progression of chronic myeloid leukemia. Leuk. Lymphoma 45, 1775–1781 (2004).
Brummendorf, T. H. et al. Prognostic implications of differences in telomere length between normal and malignant cells from patients with chronic myeloid leukemia measured by flow cytometry. Blood 95, 1883–1890 (2000). The authors show, for the first time, that telomere length has prognostic significance for CML.
Kim, N. W. et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994).
Campbell, L. J. et al. hTERT, the catalytic component of telomerase, is downregulated in the haematopoietic stem cells of patients with chronic myeloid leukaemia. Leukemia 20, 671–679 (2006).
Kharbanda, S. et al. Regulation of the hTERT telomerase catalytic subunit by the c--Abl tyrosine kinase. Curr. Biol. 10, 568–575 (2000).
Neviani, P. et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell 8, 355–368 (2005). Important evidence that antagonism of a tumour suppressor by BCR-ABL correlates with (and may be a prerequisite for) disease progression in CML.
Druker, B. J. et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 355, 2408–2417 (2006). Comprehensive follow-up study of patients on the first phase-3 clinical trial of imatinib as front-line therapy for newly diagnosed CML.
Apperley, J. F. Managing the patient with chronic myeloid leukemia through and after allogeneic stem cell transplantation. Hematology. Am. Soc. Hematol. Educ. Program. 226–232 (2006).
Kantarjian, H. et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N. Engl. J. Med. 354, 2542–2551 (2006).
Talpaz, M. et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 354, 2531–2541 (2006).
Hughes, T. & Branford, S. in Myeloproliferative disorders (eds Melo, J. V. & Goldman, J. M.) 143–164 (Springer-Verlag, Heidelberg, Germany, 2006).
Hughes, T. P. et al. Frequency of major molecular responses to imatinib or interferon α plus cytarabine in newly diagnosed chronic myeloid leukemia. N. Engl. J. Med. 349, 1423–1432 (2003).
Radich, J. P. et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc. Natl Acad. Sci. USA 103, 2794–2799 (2006). An extensive gene-expression profiling study of patients with CML, revealing gene signatures consistent with a two-step, rather than a three-step, process of disease progression.
Ohmine, K. et al. Characterization of stage progression in chronic myeloid leukemia by DNA microarray with purified hematopoietic stem cells. Oncogene 20, 8249–8257 (2001).
Ilaria, R. L., Jr. & Van Etten, R. A. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 271, 31704–31710 (1996).
Carlesso, N., Frank, D. A. & Griffin, J. D. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J. Exp. Med. 183, 811–820 (1996).
Nowicki, M. O. et al. Chronic myelogenous leukemia molecular signature. Oncogene 22, 3952–3963 (2003).
van de Vijver, M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002).
Paik, S. et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 24, 3726–3734 (2006).
Couzin, J. Diagnostics. Amid debate, gene-based cancer test approved. Science 315, 924 (2007).
Kaklamani, V. A genetic signature can predict prognosis and response to therapy in breast cancer: oncotype DX. Expert. Rev. Mol. Diagn. 6, 803–809 (2006).
Vecchi, M. et al. Gene expression analysis of early and advanced gastric cancers. Oncogene 5 February 2007 (doi: 10.1038/sj.onc.1210208).
Yong, A. S., Szydlo, R. M., Goldman, J. M., Apperley, J. F. & Melo, J. V. Molecular profiling of CD34+ cells identifies low expression of CD7, along with high expression of proteinase 3 or elastase, as predictors of longer survival in patients with CML. Blood 107, 205–212 (2006). An important and elegant gene-expression profiling study identifying biomarkers of survival in CML patients with disease stratified as either 'indolent' or 'aggressive'.
Ogata, K. et al. Reappraisal of the clinical significance of CD7 expression in association with cytogenetics in de novo acute myeloid leukaemia. Br. J. Haematol. 115, 612–615 (2001).
Ogata, K. et al. Clinical significance of phenotypic features of blasts in patients with myelodysplastic syndrome. Blood 100, 3887–3896 (2002).
Molldrem, J. J. et al. Cytotoxic T lymphocytes specific for a nonpolymorphic proteinase 3 peptide preferentially inhibit chronic myeloid leukemia colony-forming units. Blood 90, 2529–2534 (1997).
Molldrem, J. J., Lee, P. P., Wang, C., Champlin, R. E. & Davis, M. M. A PR1-human leukocyte antigen-A2 tetramer can be used to isolate low-frequency cytotoxic T lymphocytes from healthy donors that selectively lyse chronic myelogenous leukemia. Cancer Res. 59, 2675–2681 (1999).
Fujiwara, H. et al. Identification and in vitro expansion of CD4+ and CD8+ T cells specific for human neutrophil elastase. Blood 103, 3076–3083 (2004).
Mohty, M., Yong, A. S., Szydlo, R. M., Apperley, J. F. & Melo, J. V. The polycomb group BMI1 gene is a molecular marker for predicting prognosis of chronic myeloid leukemia. Blood 14 March 2007 (doi: 10.1182/blood-2006–12–065599).
Grade, M. et al. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res. 67, 41–56 (2007).
Gudmundsdottir, K. & Ashworth, A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 25, 5864–5874 (2006).
Santhanam, R. et al. FTY720, a new and alternative strategy for treating blast crisis CML and Ph1 ALL patients. Blood 108, 288 (2006).
Collis, S. J., DeWeese, T. L., Jeggo, P. A. & Parker, A. R. The life and death of DNA-PK. Oncogene 24, 949–961 (2005).
Kunkel, T. A. Considering the cancer consequences of altered DNA polymerase function. Cancer Cell 3, 105–110 (2003). An essential review of DNA polymerases and how their varied error rates have consequences for cancer.
Ahuja, H., Bar-Eli, M., Advani, S. H., Benchimol, S. & Cline, M. J. Alterations in the p53 gene and the clonal evolution of the blast crisis of chronic myelocytic leukemia. Proc. Natl Acad. Sci. USA 86, 6783–6787 (1989).
Feinstein, E. et al. p53 in chronic myelogenous leukemia in acute phase. Proc. Natl Acad. Sci. USA 88, 6293–6297 (1991).
Goetz, A. W., van der, K. H., Maya, R., Oren, M. & Aulitzky, W. E. Requirement for Mdm2 in the survival effects of Bcr-Abl and interleukin 3 in hematopoietic cells. Cancer Res. 61, 7635–7641 (2001).
Trotta, R. et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell 3, 145–160 (2003).
Towatari, M., Adachi, K., Kato, H. & Saito, H. Absence of the human retinoblastoma gene product in the megakaryoblastic crisis of chronic myelogenous leukemia. Blood 78, 2178–2181 (1991).
Ahuja, H. G., Jat, P. S., Foti, A., Bar Eli, M. & Cline, M. J. Abnormalities of the retinoblastoma gene in the pathogenesis of acute leukemia. Blood 78, 3259–3268 (1991).
Beck, Z. et al. Alterations of P53 and RB genes and the evolution of the accelerated phase of chronic myeloid leukemia. Leuk. Lymphoma 38, 587–597 (2000).
Sill, H., Goldman, J. M. & Cross, N. C. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood 85, 2013–2016 (1995).
Serra, A., Gottardi, E., Della, R. F., Saglio, G. & Iolascon, A. Involvement of the cyclin-dependent kinase-4 inhibitor (CDKN2) gene in the pathogenesis of lymphoid blast crisis of chronic myelogenous leukaemia. Br. J. Haematol. 91, 625–629 (1995).
Hernandez-Boluda, J. C. et al. Genomic p16 abnormalities in the progression of chronic myeloid leukemia into blast crisis: a sequential study in 42 patients. Exp. Hematol. 31, 204–210 (2003).
Serrano, M., Hannon, G. J. & Beach, D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366, 704–707 (1993).
Stott, F. J. et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17, 5001–5014 (1998).
Gil, J. & Peters, G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nature Rev. Mol. Cell Biol. 7, 667–677 (2006).
Sharpless, N. E. & DePinho, R. A. The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev. 9, 22–30 (1999).
Kim, W. Y. & Sharpless, N. E. The regulation of INK4/ARF in cancer and aging. Cell 127, 265–275 (2006).
Pomerantz, J. et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92, 713–723 (1998).
Nagy, E. et al. Frequent methylation of p16INK4A and p14ARF genes implicated in the evolution of chronic myeloid leukaemia from its chronic to accelerated phase. Eur. J. Cancer 39, 2298–2305 (2003).
Michor, F. Chronic myeloid leukemia blast crisis arises from progenitors. Stem Cells 25, 1114–1118 (2007).
Graham, S. M. et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood 99, 319–325 (2002).
Holtz, M. S. et al. Imatinib mesylate (STI571) inhibits growth of primitive malignant progenitors in chronic myelogenous leukemia through reversal of abnormally increased proliferation. Blood 99, 3792–3800 (2002).
Copland, M. et al. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood 107, 4532–4539 (2006).
Griswold, I. J. et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol. Cell Biol. 26, 6082–6093 (2006).
Michor, F. et al. Dynamics of chronic myeloid leukaemia. Nature 435, 1267–1270 (2005).
Stoklosa, T., Slupianek, A., Basak, G. & Skorski, T. BCR/ABL kinase disrupts formation of mismatch repair complex to induce genomic instability. Blood 106, 803A (2005).
Drummond, M. W. et al. Dysregulated expression of the major telomerase components in leukaemic stem cells. Leukemia 19, 381–389 (2005).
Acknowledgements
We are very grateful to J. M. Goldman for kindly reviewing and commenting on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
National Cancer Institute
FURTHER INFORMATION
Glossary
- Chronic phase
-
The initial stage of CML, in which most patients are diagnosed. It usually has an insidious onset, and the main clinical findings include enlarged spleen, fatigue and weight loss. The peripheral blood shows leukocytosis (approximately 150 ×109/L white blood cells (WBCs)), predominantly owing to neutrophils in different stages of maturation, as well as basophilia and eosinophilia. Blasts usually represent <2% of the WBCs. The platelet count is normal or increased.
- Accelerated phase
-
An intermediate stage of CML evolution, when the disease starts to become refractory to therapy. It is characterized by an increase in spleen size and in total WBCs, blasts comprising 10–19% of the WBCs, >20% circulating basophils, persistent thrombocytopenia and/or the appearance of new clonal cytogenetic abnormalities.
- Blast crisis
-
The final stage of CML, which may or may not be preceded by an 'accelerated phase'. Patients experience worsened performance status, and symptoms related to thrombocytopenia, anaemia and increased spleen enlargement. The WHO (World Health Organization) criteria for the diagnosis of blast crisis include: blasts in excess of 20% in the peripheral blood or bone marrow; and/or extramedullary blast proliferation; and/or large foci or clusters of blasts in bone marrow histological sections.
- Mutator phenotype
-
A mutator phenotype arises owing to mutations in genes that are crucial for maintaining genomic stability.
- Telomere
-
The termini of eukaryotic chromosomes that function to prevent the loss of genetic material. Human telomeres consist of tandem repeats of the DNA sequence TTAGGG. Telomerase is a ribonucleoprotein, required for the maintenance of telomeres, consisting of an RNA template and a catalytic subunit (TERT in humans). Whereas human stem cells express telomerase and retain intact telomeres, most somatic cells do not, and consequently have telomeres that become shorter with successive cell divisions.
- STAT5
-
A transcription factor that is activated by multiple haematopoietic cytokines. In cell lines, BCR-ABL was shown to catalyse the phosphorylation of tyrosine residues on STAT5, leading to its constitutive activation. STAT5 promotes cell survival and proliferation by transactivating anti-apoptotic genes (BCL-XL) and genes associated with cell-cycle progression (cyclin D1).
- CD34
-
A cell-surface protein expressed by haematopoietic stem cells, haematopoietic progenitor cells and endothelial cells, which is used as a marker to isolate human haematopoietic progenitor cells.
- Ataxia telangiectasia
-
A recessive disorder caused by inactivating mutations in the ATM gene and characterized by X-ray hypersensitivity, genomic instability and a predisposition to both solid tumours and haematological malignancies.
- Intra-S-phase cell-cycle checkpoint
-
Normally, the activated intra-S-phase checkpoint delays the cell cycle, providing additional time for DNA repair, or for initiating apoptosis, if the damage is irreparable.
- BRCA1-associated genome surveillance complex
-
(BASC). This DNA damage repair complex includes repair proteins and tumour suppressors (MSH2, MSH6, MLH1, ATM and BLM) as well as the RAD50–MRE11–NBS1 complex.
- Mononuclear cells
-
The fraction of leukocytes consisting of lymphocytes, monocytes and immature granulocytes (blasts, promyelocytes, myelocytes and metamyelocytes) that is obtained by density centrifugation of the peripheral blood for exclusion of the polymorphonuclear cells (that is, band and segmented granulocytes).
- Xeroderma pigmentosum
-
(XP). An inherited genetic disorder characterized by an inability to repair DNA damage resulting from exposure to UV light. Before the discovery of the mutated genes that cause this condition, patients were classified into 7 complementation groups (XPA, XPB, XPC, XPD, XPE, XPF and XPG).
- Depurination
-
A form of spontaneous DNA damage in which purines (adenine or guanine) are lost from a DNA strand resulting in the formation of an 'abasic' site.
- Deamination
-
Another form of spontaneous DNA damage in which amino groups are lost from the bases adenine, guanine, cytosine or 5-methylcytosine, converting them to their miscoding equivalents of hypoxanthine, xanthine, uracil and thymine, respectively.
- Rubinstein–Taybi malformation syndrome
-
An inherited condition characterized by short stature, broad thumbs and first toes, unusual facial features and moderate to severe mental retardation. Individuals with this condition are predisposed to developing non-malignant and malignant tumours, as well as having an increased risk of developing leukaemia or lymphoma. The condition is caused by inactivating autosomal dominant mutations of the CREBBP gene, which is involved in regulating cell division and growth.
- Multivariate analysis
-
The application of a statistical method for the simultaneous analysis of more than two variables.
- Primary azurophilic granules
-
Lysosomes found in the cytoplasm of granulocytes that store enzymes, essential for catalysing the 'respiratory burst' of neutrophils that is directed against pathogens or infected cells. The lysosomes are known as 'azurophilic' granules as they are readily stained with azure dyes.
Rights and permissions
About this article
Cite this article
Melo, J., Barnes, D. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer 7, 441–453 (2007). https://doi.org/10.1038/nrc2147
Issue Date:
DOI: https://doi.org/10.1038/nrc2147
This article is cited by
-
The importance of personalized medicine in chronic myeloid leukemia management: a narrative review
Egyptian Journal of Medical Human Genetics (2023)
-
BCR-ABL triggers a glucose-dependent survival program during leukemogenesis through the suppression of TXNIP
Cell Death & Disease (2023)
-
EVI1 upregulates PTGS1 (COX1) and decreases the action of tyrosine kinase inhibitors (TKIs) in chronic myeloid leukemia cells
International Journal of Hematology (2023)
-
Genotoxicity assessment: opportunities, challenges and perspectives for quantitative evaluations of dose–response data
Archives of Toxicology (2023)
-
Combination venetoclax and olverembatinib (HQP1351) as a successful therapeutic strategy for relapsed/refractory (R/R) mixed-phenotype blast phase of chronic myeloid leukemia
Annals of Hematology (2023)