Key Points
-
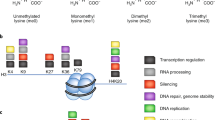
Lysine methylation is widely recognized as a fundamental post-translational modification.
-
Most protein lysine methyltransferases (PKMTs) contain the SET domain, but several non-SET proteins such as DOT1-like histone H3 lysine 79 methyltransferase (DOT1L), methyltransferase-like 10 (METTL10) and METTL21A are also known to have lysine N-methyltransferase activity.
-
Protein lysine demethylases (PKDMs) consist of the lysine-specific demethylase 1 (LSD1) family, which are flavin-dependent monoamine oxidases, and the Jumonji C (JmjC) domain-containing proteins, which are α-ketoglutarate-dependent Fe(ii) dioxygenases.
-
The biological importance of protein lysine methylation in cancer can be categorized into five different functions: effect on other protein modifications; protein–protein interactions; protein stability; subcellular localization; and promoter binding.
-
Although the most widely recognized function of PKMTs and PKDMs in cancer is their effects on histones and epigenetic regulation, nearly 20 non-histone proteins related to human cancer, including p53 and RB1, have also been discovered to be methylated at lysine residues.
-
Somatic mutations of PKMTs and PKDMs are frequently found in human cancer, and it is possible that they may affect the methylation of non-histone substrates.
-
Inhibitors targeting PKMTs and PKDMs are considered to be promising agents for anticancer therapy, and it is important that a thorough understanding of all of the substrates of these enzymes is made to discern the precise mechanism of action of these inhibitors.
Abstract
Several protein lysine methyltransferases and demethylases have been identified to have critical roles in histone modification. A large body of evidence has indicated that their dysregulation is involved in the development and progression of various diseases, including cancer, and these enzymes are now considered to be potential therapeutic targets. Although most studies have focused on histone methylation, many reports have revealed that these enzymes also regulate the methylation dynamics of non-histone proteins such as p53, RB1 and STAT3 (signal transducer and activator of transcription 3), which have important roles in human tumorigenesis. In this Review, we summarize the molecular functions of protein lysine methylation and its involvement in human cancer, with a particular focus on lysine methylation of non-histone proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ambler, R. P. & Rees, M. W. ε-N-methyl-lysine in bacterial flagellar protein. Nature 184, 56–57 (1959).
Murray, K. The occurrence of ε-N-methyl lysine in histones. Biochemistry 3, 10–15 (1964).
Clarke, S. G. & Tamanoi, F. (eds) The Enzymes: Protein Methyltransferases Vol. 24 (Academic Press, 2006).
Hamamoto, R. et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nature Cell Biol. 6, 731–740 (2004). This is the pioneering report to describe that dysregulation of a PKMT can cause tumorigenesis.
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000). This is the first report to show the existence of a PKMT.
Tachibana, M., Sugimoto, K., Fukushima, T. & Shinkai, Y. SET domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276, 25309–25317 (2001).
Schlichter, A. & Cairns, B. R. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J. 24, 1222–1231 (2005).
Ringrose, L., Ehret, H. & Paro, R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell 16, 641–653 (2004).
Feng, Q. et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 (2002).
Bannister, A. J., Schneider, R. & Kouzarides, T. Histone methylation: dynamic or static? Cell 109, 801–806 (2002).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). This is the first report to show the existence of a PKDM.
Tsukada, Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006).
Hamamoto, R. et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 97, 113–118 (2006).
Kotake, Y. et al. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4α tumor suppressor gene. Genes Dev. 21, 49–54 (2007).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012). This study describes the importance of somatic mutations in PKMTs in developing anticancer drugs.
Fiskus, W. et al. Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia 28, 2155–2164 (2014).
Takawa, M. Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer Sci. 102, 1298–1305 (2011).
Sasaki, M., Yamaguchi, J., Itatsu, K., Ikeda, H. & Nakanuma, Y. Over-expression of polycomb group protein EZH2 relates to decreased expression of p16INK4a in cholangiocarcinogenesis in hepatolithiasis. J. Pathol. 215, 175–183 (2008).
Cho, H. S. et al. Enhanced expression of EHMT2 is involved in the proliferation of cancer cells through negative regulation of SIAH1. Neoplasia 13, 676–684 (2011).
Cho, H. S. et al. The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int. J. Cancer 131, E179–189 (2012).
Toyokawa, G. et al. The histone demethylase JMJD2B plays an essential role in human carcinogenesis through positive regulation of cyclin-dependent kinase 6. Cancer Prev. Res. (Phila) 4, 2051–2061 (2011).
Cho, H. S. et al. RB1 methylation by SMYD2 enhances cell cycle progression through an Increase of RB1 phosphorylation. Neoplasia 14, 476–486 (2012).
Cho, H. S. et al. Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nature Commun. 3, 1072 (2012). This study provides clear evidence that protein lysine methylation plays a critical part in the regulation of subcellular localization.
Cho, H. S. et al. Demethylation of RB regulator MYPT1 by histone demethylase LSD1 promotes cell cycle progression in cancer cells. Cancer Res. 71, 1–6 (2011).
Hamamoto, R., Toyokawa, G., Nakakido, M., Ueda, K. & Nakamura, Y. SMYD2-dependent HSP90 methylation promotes cancer cell proliferation by regulating the chaperone complex formation. Cancer Lett. 351, 126–133 (2014).
Kunizaki, M. et al. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 67, 10759–10765 (2007).
Piao, L. et al. The histone methyltransferase SMYD2 methylates PARP1 and promotes poly(ADP-ribosyl)ation activity in cancer cells. Neoplasia 16, 257–264.e2 (2014).
Smith, B. C. & Denu, J. M. Chemical mechanisms of histone lysine and arginine modifications. Biochim. Biophys. Acta 1789, 45–57 (2008).
Shimazu, T., Barjau, J., Sohtome, Y., Sodeoka, M. & Shinkai, Y. Selenium-based S-adenosylmethionine analog reveals the mammalian seven-β-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. PLoS ONE 9, e105394 (2014).
Fang, R. et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell 39, 222–233 (2010).
Kogure, M. et al. The oncogenic polycomb histone methyltransferase EZH2 methylates lysine 120 on histone H2B and competes ubiquitination. Neoplasia 15, 1251–1261 (2013).
Sone, K. et al. Critical role of lysine 134 methylation on histone H2AX for γ-H2AX production & DNA repair. Nature Commun 5, 5691 (2014).
Mazur, P. K. et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510, 283–287 (2014). This study provides strong evidence that a PKMT regulates the function of a cytoplasmic protein.
Khorasanizadeh, S. Recognition of methylated histones: new twists and variations. Curr. Opin. Struct. Biol. 21, 744–749 (2011).
Daze, K. D. & Hof, F. The cation-pi interaction at protein–protein interaction interfaces: developing and learning from synthetic mimics of proteins that bind methylated lysines. Acc. Chem. Res. 46, 937–945 (2013).
Pang, C. N., Gasteiger, E. & Wilkins, M. R. Identification of arginine- and lysine-methylation in the proteome of Saccharomyces cerevisiae and its functional implications. BMC Genomics 11, 92 (2010). This study comprehensively analyses lysine methylation of yeast proteins and identifies numerous substrates.
Lee, J. M. et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell 48, 572–586 (2012).
Hodel, M. R., Corbett, A. H. & Hodel, A. E. Dissection of a nuclear localization signal. J. Biol. Chem. 276, 1317–1325 (2001).
Chuikov, S. et al. Regulation of p53 activity through lysine methylation. Nature 432, 353–360 (2004). This paper provides the first evidence that p53 is a substrate of a PKMT.
Stark, G. R., Wang, Y. & Lu, T. Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res. 21, 375–380 (2011).
Lu, T. et al. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl Acad. Sci. USA 107, 46–51 (2010).
Lu, T. et al. Role of lysine methylation of NF-κB in differential gene regulation. Proc. Natl Acad. Sci. USA 110, 13510–13515 (2013).
Nakamura, Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 95, 7–11 (2004).
Huang, J. et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 444, 629–632 (2006).
Huang, J. et al. p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007). This study provides the first evidence that a PKDM can also demethylate a non-histone protein.
Hayami, S. et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int. J. Cancer 128, 574–586 (2011).
Shi, X. et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell 27, 636–646 (2007).
Goodrich, D. W. The retinoblastoma tumor-suppressor gene, the exception that proves the rule. Oncogene 25, 5233–5243 (2006).
Munger, K. et al. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8, 4099–4105 (1989).
Buchkovich, K., Duffy, L. A. & Harlow, E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell 58, 1097–1105 (1989).
Saddic, L. A. et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 285, 37733–37740 (2010).
Carr, S. M., Munro, S., Kessler, B., Oppermann, U. & La Thangue, N. B. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J. 30, 317–327 (2011).
Tsantoulis, P. K. & Gorgoulis, V. G. Involvement of E2F transcription factor family in cancer. Eur. J. Cancer 41, 2403–2414 (2005).
Chen, H. Z., Tsai, S. Y. & Leone, G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature Rev. Cancer 9, 785–797 (2009).
Kontaki, H. & Talianidis, I. Lysine methylation regulates E2F1-induced cell death. Mol. Cell 39, 152–160 (2010).
Ciocca, D. R. & Calderwood, S. K. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10, 86–103 (2005).
Trepel, J., Mollapour, M., Giaccone, G. & Neckers, L. Targeting the dynamic HSP90 complex in cancer. Nature Rev. Cancer 10, 537–549 (2010).
Kampinga, H. H. & Craig, E. A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nature Rev. Mol. Cell Biol. 11, 579–592 (2010).
Wandinger, S. K., Richter, K. & Buchner, J. The Hsp90 chaperone machinery. J. Biol. Chem. 283, 18473–18477 (2008).
Pratt, W. B. & Toft, D. O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228, 111–133 (2003).
Whitesell, L. & Lindquist, S. L. HSP90 and the chaperoning of cancer. Nature Rev. Cancer 5, 761–772 (2005).
Olsson, A. K., Dimberg, A., Kreuger, J. & Claesson-Welsh, L. VEGF receptor signalling — in control of vascular function. Nature Rev. Mol. Cell Biol. 7, 359–371 (2006).
Fan, F. et al. Expression and function of vascular endothelial growth factor receptor-1 on human colorectal cancer cells. Oncogene 24, 2647–2653 (2005).
Abdelrahim, M. et al. Regulation of vascular endothelial growth factor receptor-1 expression by specificity proteins 1, 3, and 4 in pancreatic cancer cells. Cancer Res. 67, 3286–3294 (2007).
Silverman, N. & Maniatis, T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15, 2321–2342 (2001).
Ea, C. K. & Baltimore, D. Regulation of NF-κB activity through lysine monomethylation of p65. Proc. Natl Acad. Sci. USA 106, 18972–18977 (2009).
Levy, D. et al. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nature Immunol. 12, 29–36 (2011).
Ali, S. & Coombes, R. C. Estrogen receptor α in human breast cancer: occurrence and significance. J. Mammary Gland Biol. Neoplasia 5, 271–281 (2000).
Zhang, X. et al. Regulation of estrogen receptor α by histone methyltransferase SMYD2-mediated protein methylation. Proc. Natl Acad. Sci. USA 110, 17284–17289 (2013).
Grimm, S. L. & Rosen, J. M. The role of C/EBPβ in mammary gland development and breast cancer. J. Mammary Gland Biol. Neoplasia 8, 191–204 (2003).
Pless, O. et al. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-β. J. Biol. Chem. 283, 26357–26363 (2008).
Chen, Y. et al. STAT3, a poor survival predicator, is associated with lymph node metastasis from breast cancer. J. Breast Cancer 16, 40–49 (2013).
Yang, J. et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl Acad. Sci. USA 107, 21499–21504 (2010).
Kim, E. et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 23, 839–852 (2013). This study provides clear evidence that non-histone lysine methylation is an important target of cancer therapy in vivo.
Takawa, M. et al. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 72, 3217–3227 (2012).
Zhao, H. et al. Targeting tyrosine phosphorylation of PCNA inhibits prostate cancer growth. Mol. Cancer Ther. 10, 29–36 (2011).
Chen, A. PARP inhibitors: its role in treatment of cancer. Chin. J. Cancer 30, 463–471 (2011).
Jaffe, J. D. et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nature Genet. 45, 1386–1391 (2013). This study describes the importance of somatic mutations in PKMTs for developing anticancer drugs.
Yin, S. et al. Exome sequencing identifies frequent mutation of MLL2 in non-small cell lung carcinoma from Chinese patients. Sci. Rep. 4, 6036 (2014).
Gossage, L. et al. Clinical and pathological impact of VHL, PBRM1, BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma. Genes Chromosomes Cancer 53, 38–51 (2014).
Cleary, S. P. et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 58, 1693–1702 (2013).
Dubuc, A. M. et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 125, 373–384 (2013).
Jankowska, A. M. et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood 118, 3932–3941 (2011).
Morin, R. D. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Wang, X. X. et al. Somatic mutations of the mixed-lineage leukemia 3 (MLL3) gene in primary breast cancers. Pathol. Oncol. Res. 17, 429–433 (2011).
Gui, Y. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nature Genet. 43, 875–878 (2011).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Lindberg, J. et al. The mitochondrial and autosomal mutation landscapes of prostate cancer. Eur. Urol. 63, 702–708 (2013).
Ho, A. S. et al. The mutational landscape of adenoid cystic carcinoma. Nature Genet. 45, 791–798 (2013).
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013).
Greiner, D., Bonaldi, T., Eskeland, R., Roemer, E. & Imhof, A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nature Chem. Biol. 1, 143–145 (2005).
Kubicek, S. et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481 (2007).
Chang, Y. et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nature Struct. Mol. Biol. 16, 312–317 (2009). This study determined the crystal structure of the catalytic SET domain of G9a-like protein in complex with the specific inhibitor BIX-01294.
Liu, Y. & Gray, N. S. Rational design of inhibitors that bind to inactive kinase conformations. Nature Chem. Biol. 2, 358–364 (2006).
Vedadi, M. et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nature Chem. Biol. 7, 566–574 (2011).
Ferguson, A. D. et al. Structural basis of substrate methylation and inhibition of SMYD2. Structure 19, 1262–1273 (2011). This study describes a drug development strategy for inhibitors of a PKMT using a non-histone protein as a substrate.
Knutson, S. K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature Chem. Biol. 8, 890–896 (2012).
Knutson, S. K. et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13, 842–854 (2014).
Nikoloski, G. et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature Genet. 42, 665–667 (2010).
Bejar, R. et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 30, 3376–3382 (2012).
Jerez, A. et al. Loss of heterozygosity in 7q myeloid disorders: clinical associations and genomic pathogenesis. Blood 119, 6109–6117 (2012).
Muto, T. et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J. Exp. Med. 210, 2627–2639 (2013).
Basavapathruni, A. et al. Nonclinical pharmacokinetics and metabolism of EPZ-5676, a novel DOT1L histone methyltransferase inhibitor. Biopharm. Drug Dispos. 35, 237–252 (2014).
Klaus, C. R. et al. DOT1L inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs and hypomethylating agents in MLL-rearranged leukemia cells. J. Pharmacol. Exp. Ther. 350, 646–656 (2014). References 103 and 104 describe the DOT1L inhibitor EPZ5676, which is currently in clinical trials.
Schmidt, D. M. & McCafferty, D. G. Trans-2-phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 46, 4408–4416 (2007).
Schenk, T. et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nature Med. 18, 605–611 (2012).
Wang, J. et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 71, 7238–7249 (2011).
Kruidenier, L. et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408 (2012). This report provides evidence that a Jumonji-type demethylase is a good target for drug discovery.
Wang, L. et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nature Commun. 4, 2035 (2013).
Singh, B. N. et al. Nonhistone protein acetylation as cancer therapy targets. Expert Rev. Anticancer Ther. 10, 935–954 (2010).
Komatsu, S. et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis 30, 1139–1146 (2009).
Abu-Farha, M. et al. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol. Cell Proteom. 7, 560–572 (2008).
Brown, M. A., Sims, R. J., 3rd, Gottlieb, P. D. & Tucker, P. W. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 5, 26 (2006).
Silva, F. P. et al. Enhanced methyltransferase activity of SMYD3 by the cleavage of its N-terminal region in human cancer cells. Oncogene 27, 2686–2692 (2008).
Foreman, K. W. et al. Structural and functional profiling of the human histone methyltransferase SMYD3. PLoS ONE 6, e22290 (2011).
Dong, S. W. et al. Effect of the downregulation of SMYD3 expression by RNAi on RIZ1 expression and proliferation of esophageal squamous cell carcinoma. Oncol. Rep. 32, 1064–1070 (2014).
Sponziello, M. et al. Epigenetic-related gene expression profile in medullary thyroid cancer revealed the overexpression of the histone methyltransferases EZH2 and SMYD3 in aggressive tumours. Mol. Cell Endocrinol. 392, 8–13 (2014).
Vieira, F. Q. et al. Deregulated expression of selected histone methylases and demethylases in prostate carcinoma. Endocr. Relat. Cancer 21, 51–61 (2014).
Wang, S. Z. et al. Knockdown of SMYD3 by RNA interference inhibits cervical carcinoma cell growth and invasion in vitro. BMB Rep. 41, 294–299 (2008).
Zeng, B. et al. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 586, 3271–3278 (2012).
Guo, N. et al. Hepatitis C virus core upregulates the methylation status of the RASSF1A promoter through regulation of SMYD3 in hilar cholangiocarcinoma cells. Acta Biochim. Biophys. Sin. (Shanghai) 43, 354–361 (2011).
Wang, G. G., Cai, L., Pasillas, M. P. & Kamps, M. P. NUP98–NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nature Cell Biol. 9, 804–812 (2007).
Panarello, C., Rosanda, C. & Morerio, C. Cryptic translocation t(5;11)(q35;p15.5) with involvement of the NSD1 and NUP98 genes without 5q deletion in childhood acute myeloid leukemia. Genes Chromosomes Cancer 35, 277–281 (2002).
Hollink, I. H. et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood 118, 3645–3656 (2011).
Toyokawa, G. et al. Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia 13, 887–898 (2011).
Martinez-Garcia, E. et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 117, 211–220 (2011).
Ezponda, T. et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene 32, 2882–2890 (2013).
Kim, J. Y. et al. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol. Cell. Biol. 28, 2023–2034 (2008).
Pei, H. et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 (2011).
Kang, D. et al. The histone methyltransferase Wolf–Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes Chromosomes Cancer 52, 126–139 (2013).
Zhou, Z., Thomsen, R., Kahns, S. & Nielsen, A. L. The NSD3L histone methyltransferase regulates cell cycle and cell invasion in breast cancer cells. Biochem. Biophys. Res. Commun. 398, 565–570 (2010).
Kim, S. M. et al. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem. Biophys. Res. Commun. 345, 318–323 (2006).
Salz, T. et al. hSETD1A regulates Wnt target genes and controls tumor growth of colorectal cancer cells. Cancer Res. 74, 775–786 (2014).
Hu, H. Y. et al. Set9, NF-κB, and microRNA-21 mediate berberine-induced apoptosis of human multiple myeloma cells. Acta Pharmacol. Sin. 34, 157–166 (2013).
Subramanian, K. et al. Regulation of estrogen receptor α by the SET7 lysine methyltransferase. Mol. Cell 30, 336–347 (2008).
O'Carroll, D. et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 20, 9423–9433 (2000).
Lehnertz, B. et al. The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes Dev. 28, 317–327 (2014).
Zhong, X. et al. Overexpression of G9a and MCM7 in esophageal squamous cell carcinoma associated with poor prognosis. Histopathology http://dx.doi.org/10.1111/his.12456 (2014).
Natarajan, T. G. et al. Epigenetic regulator MLL2 shows altered expression in cancer cell lines and tumors from human breast and colon. Cancer Cell. Int. 10, 13 (2010).
Xia, M. et al. Downregulation of MLL3 in esophageal squamous cell carcinoma is required for the growth and metastasis of cancer cells. Tumour Biol. http://dx.doi.org/10.1007/s13277-014-2616-3 (2014).
Li, B. et al. Association of MLL3 expression with prognosis in gastric cancer. Genet. Mol. Res. 13, 7513–7518 (2014).
Ruault, M., Brun, M. E., Ventura, M., Roizes, G. & De Sario, A. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene 284, 73–81 (2002).
Okada, Y. et al. hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178 (2005).
Nguyen, A. T., Taranova, O., He, J. & Zhang, Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 117, 6912–6922 (2011).
Bernt, K. M. & Armstrong, S. A. A role for DOT1L in MLL-rearranged leukemias. Epigenomics 3, 667–670 (2011).
Yamada, D. et al. Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma: clinical impact on recurrence after hepatic resection. Ann. Surg. Oncol. 19, S355–S364 (2012).
Guo, X. et al. The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma 58, 153–157 (2011).
Kogure, M. et al. Deregulation of the histone demethylase JMJD2A is involved in human carcinogenesis through regulation of the G1/S transition. Cancer Lett. 336, 76–84 (2013).
Berry, W. L., Shin, S., Lightfoot, S. A. & Janknecht, R. Oncogenic features of the JMJD2A histone demethylase in breast cancer. Int. J. Oncol. 41, 1701–1706 (2012).
Wang, H. L. et al. Expression and effects of JMJD2A histone demethylase in endometrial carcinoma. Asian Pac. J. Cancer Prev. 15, 3051–3056 (2014).
Hu, C. E., Liu, Y. C., Zhang, H. D. & Huang, G. J. JMJD2A predicts prognosis and regulates cell growth in human gastric cancer. Biochem. Biophys. Res. Commun. 449, 1–7 (2014).
Sun, B. B. et al. Silencing of JMJD2B induces cell apoptosis via mitochondria-mediated and death receptor-mediated pathway activation in colorectal cancer. J. Dig. Dis. 15, 491–500 (2014).
Chen, L. et al. Jumonji domain-containing protein 2B silencing induces DNA damage response via STAT3 pathway in colorectal cancer. Br. J. Cancer 110, 1014–1026 (2014).
Zhao, L. et al. JMJD2B promotes epithelial-mesenchymal transition by cooperating with β-catenin and enhances gastric cancer metastasis. Clin. Cancer Res. 19, 6419–6429 (2013).
Ene, C. I. et al. Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS ONE 7, e51407 (2012).
Nickerson, M. L. et al. Concurrent alterations in TERT, KDM6A, and the BRCA pathway in bladder cancer. Clin. Cancer Res. 20, 4935–4948 (2014).
Ntziachristos, P. et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 514, 513–517 (2014).
Shen, Y. et al. Expression and significance of histone H3K27 demethylases in renal cell carcinoma. BMC Cancer 12, 470 (2012).
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genet. 41, 521–523 (2009).
Hayami, S. et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol. Cancer 9, 59 (2010).
Yamamoto, S. et al. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell 25, 762–777 (2014).
Kano, Y. et al. Jumonji/Arid1b (Jarid1b) protein modulates human esophageal cancer cell growth. Mol. Clin. Oncol. 1, 753–757 (2013).
Ohta, K. et al. Depletion of JARID1B induces cellular senescence in human colorectal cancer. Int. J. Oncol. 42, 1212–1218 (2013).
Radberger, P., Radberger, A., Bykov, V. J., Seregard, S. & Economou, M. A. JARID1B protein expression and prognostic implications in uveal melanoma. Invest. Ophthalmol. Vis. Sci. 53, 4442–4449 (2012).
Acknowledgements
The authors express great gratitude to the past and present members of Y.N.'s laboratory.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Y.N. is a stock holder and a scientific adviser of Oncotherapy Science. Y.N. has also received research grants from Oncotherapy Science. No potential competing interests were disclosed by R.H. and V.S.
Related links
Supplementary information
41568_2015_BFnrc3884_MOESM20_ESM.pdf
Supplementary information S1 (table) | Somatic mutations of PKMTs and PKDMs frequently found in human cancer (PDF 3573 kb)
Glossary
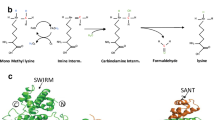
- S-adenosyl-L-methionine
-
(AdoMet). A molecule synthesized from methionine and ATP by methionine adenosyltransferase. The methylation group attached to the methionine sulphur atom in AdoMet is chemically reactive. This allows donation of this group to an acceptor substrate in transmethylation reactions.
- Monoamine oxidase
-
A family of enzymes that catalyse the oxidation of monoamines. They belong to the protein family of flavin-containing amine oxidoreductases.
- Oxygenase
-
An enzyme of the oxidoreductase class that catalyses the incorporation of both atoms of molecular oxygen into the substrate.
- Tudor domains
-
Protein domains originally identified as a region of 50 amino acids found in the Drosophila melanogaster Tudor protein. The structurally characterized Tudor domain in human proteins recognizes symmetrically dimethylated arginine. This domain is also reported as a methyl lysine-binding protein module.
- Nitrosylation
-
Nitrosylation, specifically S-nitrosylation, involves the covalent incorporation of a nitric oxide moiety into thiol groups, to form S-nitrosothiol. S-nitrosylation is a physiologically important post-translational modification that affects a variety of proteins involved in a number of cellular processes.
- Okazaki fragments
-
Short, newly synthesized DNA fragments that are formed on the lagging template strand during DNA replication. The fragments are produced because of the need for DNA polymerase to always synthesize in a 5′-to-3′ direction and are subsequently ligated together to form a continuous strand.
Rights and permissions
About this article
Cite this article
Hamamoto, R., Saloura, V. & Nakamura, Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer 15, 110–124 (2015). https://doi.org/10.1038/nrc3884
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3884
This article is cited by
-
Computational approach for assessing the involvement of SMYD2 protein in human cancers using TCGA data
Journal of Genetic Engineering and Biotechnology (2023)
-
Epigenetic modification of m6A regulator proteins in cancer
Molecular Cancer (2023)
-
PRMT5 methylating SMAD4 activates TGF-β signaling and promotes colorectal cancer metastasis
Oncogene (2023)
-
Epigenetic regulation of SMAD3 by histone methyltransferase SMYD2 promotes lung cancer metastasis
Experimental & Molecular Medicine (2023)
-
Global lysine methylome profiling using systematically characterized affinity reagents
Scientific Reports (2023)