Key Points
-
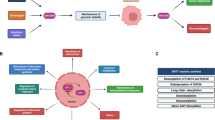
Sirtuins are NAD+-dependent enzymes that modulate the activities of target proteins, mediate changes in the metabolic status of cells and regulate diseases of ageing, including neurodegeneration, diabetes, cardiovascular diseases and cancer.
-
Sirtuin 1 (SIRT1) was the first sirtuin to be shown to be involved in cancer. It was demonstrated that SIRT1 represses p53-mediated tumour suppression. Since this finding, SIRT1 has been shown to have both tumour-suppressive and oncogenic roles, depending on the type and stage of cancer.
-
The mitochondrial sirtuin SIRT3 regulates levels of reactive oxygen species (ROS). Loss of SIRT3 results in increased levels of ROS, with subsequent activation of hypoxia-inducible factor 1α (HIF1α) and increased expression of HIF1 target genes, including glycolysis and angiogenesis genes, which favour tumour growth.
-
The mitochondrial sirtuin SIRT4 regulates glutamine metabolism by inhibiting glutamate dehydrogenase (GDH), the rate-limiting enzyme in glutaminolysis. Loss of SIRT4 leads to increased glutamine catabolism, a pathway that promotes tumour growth.
-
The chromatin-bound sirtuin SIRT6 regulates cancer metabolism and inflammation by acting as a co-repressor for HIF1α, MYC and nuclear factor-κB (NF-κB). Loss of SIRT6 results in increased expression of glycolysis, glutaminolysis and ribosomal genes, all of which favour proliferation and tumorigenesis.
-
SIRT1 and SIRT6 promote genomic stability by regulating both single- and double-strand DNA break-repair pathways.
Abstract
The sirtuins (SIRTs; of which there are seven in mammals) are NAD+-dependent enzymes that regulate a large number of cellular pathways and forestall the progression of ageing and age-associated diseases. In recent years, the role of sirtuins in cancer biology has become increasingly apparent, and growing evidence demonstrates that sirtuins regulate many processes that go awry in cancer cells, such as cellular metabolism, the regulation of chromatin structure and the maintenance of genomic stability. In this article, we review recent advances in our understanding of how sirtuins affect cancer metabolism, DNA repair and the tumour microenvironment and how activating or inhibiting sirtuins may be important in preventing or treating cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Imai, S., Armstrong, C. M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000). This study shows that yeast Sir2 and the mouse homologue are NAD+-dependent histone deacetylases that deacetylate histone H3K9, H3K14 and H4K16. This study also shows that Sir2 deacetylase activity is responsible for silencing, suppression of recombination and lifespan extension in vivo.
Guarente, L. Calorie restriction and sirtuins revisited. Genes Dev. 27, 2072–2085 (2013).
Guarente, L. Franklin, H. Epstein lecture: sirtuins, aging, and medicine. N. Engl. J. Med. 364, 2235–2244 (2011).
Frye, R. A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 (2000).
Ward, P. S. & Thompson, C. B. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 (2012).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Chalkiadaki, A. & Guarente, L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 8, 287–296 (2012).
Chang, H. C. & Guarente, L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 25, 138–145 (2014).
Lagouge, M. et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127, 1109–1122 (2006).
Milne, J. C. et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716 (2007).
Baur, J. A. et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342 (2006).
Feige, J. N. et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell. Metab. 8, 347–358 (2008).
Bordone, L. et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6, 759–767 (2007).
Herranz, D. et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat. Commun. 1, 3 (2010).
Pfluger, P. T., Herranz, D., Velasco-Miguel, S., Serrano, M. & Tschop, M. H. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl Acad. Sci. USA 105, 9793–9798 (2008).
Luo, J. et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148 (2001).
Vaziri, H. et al. hSIR2 SIRT1 functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159 (2001). References 16 and 17 show that, on DNA damage, SIRT1 binds and deacetylates p53, resulting in inhibition of its transcriptional activity and subsequent cellular events: namely, growth arrest and apoptosis.
Cheng, H. L. et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl Acad. Sci. USA 100, 10794–10799 (2003). This paper demonstrates that Sirt1 -knockout mice exhibit developmental defects. After DNA damage, Sirt1 -knockout fibroblasts have hyperacetylated p53 and are resistant to apoptosis.
Wang, C. et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 8, 1025–1031 (2006).
Firestein, R. et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE 3, e2020 (2008).
Oberdoerffer, P. et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907–918 (2008). This paper shows that, on DNA damage, SIRT1 is relocalized from gene targets to DNA breaks to promote repair and genomic stability.
Wang, R. H. et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14, 312–323 (2008). This article reveals that reduced levels of SIRT1 in Trp53 -heterozygous mice lead to tumorigenesis in many tissues.
Chen, I. C. et al. Role of SIRT1 in regulation of epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis. Mol. Cancer 13, 254 (2014).
Wang, R. H. et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol. Cell 32, 11–20 (2008).
Simic, P. et al. SIRT1 suppresses the epithelial-to-mesenchymal transition in cancer metastasis and organ fibrosis. Cell Rep. 3, 1175–1186 (2013).
Tiberi, L. et al. A BCL6/BCOR/SIRT1 complex triggers neurogenesis and suppresses medulloblastoma by repressing Sonic Hedgehog signaling. Cancer Cell 26, 797–812 (2014).
Chauhan, D. et al. Preclinical evaluation of a novel SIRT1 modulator SRT1720 in multiple myeloma cells. Br. J. Haematol. 155, 588–598 (2011).
Lahusen, T. J. & Deng, C. X. SRT1720 induces lysosomal-dependent cell death of breast cancer cells. Mol. Cancer Ther. 14, 183–192 (2015).
Chen, C. W. et al. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat. Med. 21, 335–343 (2015).
Jang, K. Y. et al. SIRT1 and c-Myc promote liver tumor cell survival and predict poor survival of human hepatocellular carcinomas. PLoS ONE 7, e45119 (2012).
Chen, X. et al. High levels of SIRT1 expression enhance tumorigenesis and associate with a poor prognosis of colorectal carcinoma patients. Sci. Rep. 4, 7481 (2014).
Menssen, A. et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc. Natl Acad. Sci. USA 109, E187–E196 (2012).
Li, L. et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell 15, 431–446 (2014).
Li, L. et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell 21, 266–281 (2012). This paper describes how a combination of SIRT1 inhibition and BCR–ABL inhibition in leukaemia stem cells led to reduced growth and increased apoptosis.
Herranz, D. et al. SIRT1 promotes thyroid carcinogenesis driven by PTEN deficiency. Oncogene 32, 4052–4056 (2013).
Boily, G., He, X. H., Pearce, B., Jardine, K. & McBurney, M. W. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene 28, 2882–2893 (2009).
Leko, V., Park, G. J., Lao, U., Simon, J. A. & Bedalov, A. Enterocyte-specific inactivation of SIRT1 reduces tumor load in the APC+/min mouse model. PLoS ONE 8, e66283 (2013).
Chen, W. Y. et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123, 437–448 (2005).
Wales, M. M. et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat. Med. 1, 570–577 (1995).
Hamaguchi, M. et al. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc. Natl Acad. Sci. USA 99, 13647–13652 (2002).
Kim, J. E., Chen, J. & Lou, Z. DBC1 is a negative regulator of SIRT1. Nature 451, 583–586 (2008).
Zhao, W. et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587–590 (2008). References 41 and 42 demonstrate that DBC1 inhibits SIRT1 in human cells, leading to increased levels of p53 acetylation and transcriptional activity.
Nemoto, S., Fergusson, M. M. & Finkel, T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 306, 2105–2108 (2004).
Chang, T. C. et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752 (2007).
Welch, C., Chen, Y. & Stallings, R. L. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26, 5017–5022 (2007).
Yamakuchi, M., Ferlito, M. & Lowenstein, C. J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl Acad. Sci. USA 105, 13421–13426 (2008).
Yuan, H. et al. Activation of stress response gene SIRT1 by BCR–ABL promotes leukemogenesis. Blood 119, 1904–1914 (2012). This study shows that the oncoprotein BCR–ABL activates SIRT1 in haematopoietic stem cells and promotes their survival and proliferation, leading to leukaemia.
Wang, Z. et al. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene 32, 589–598 (2013).
Chu, F., Chou, P. M., Zheng, X., Mirkin, B. L. & Rebbaa, A. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Res. 65, 10183–10187 (2005).
Ota, H. et al. Sirt1 inhibitor, sirtinol, induces senescence-like growth arrest with attenuated Ras–MAPK signaling in human cancer cells. Oncogene 25, 176–185 (2006).
Heltweg, B. et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 66, 4368–4377 (2006).
Kim, H. S. et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20, 487–499 (2011). This study shows that SIRT2 regulates the cell cycle and that Sirt2 knockout leads to tumorigenesis in mice by increasing genomic instability.
Vaquero, A. et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 20, 1256–1261 (2006).
Lin, Z. Z. et al. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B overexpression in HCC. BMC Cancer 10, 461 (2010).
Nadler, Y. et al. Expression of Aurora A (but not Aurora B) is predictive of survival in breast cancer. Clin. Cancer Res. 14, 4455–4462 (2008).
North, B. J. et al. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 33, 1438–1453 (2014).
Serrano, L. et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 27, 639–653 (2013).
Kumar, S. & Lombard, D. B. Mitochondrial sirtuins and their relationships with metabolic disease and cancer. Antioxid. Redox Signal. 22, 1060–1077 (2015).
Hirschey, M. D. et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 (2010).
Someya, S. et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143, 802–812 (2010).
Kim, H. S. et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17, 41–52 (2010). This article reports that Sirt3 -knockout cells have increased superoxide levels and are prone to genomic instability and transformation, and that Sirt3 -null mice develop tumours.
Finley, L. W. et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 19, 416–428 (2011). This study shows that SIRT3 functions as a tumour suppressor by reducing ROS production and destabilizing HIF1α.
Jeong, S. M. et al. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene 34, 2115–2124 (2014).
Aury-Landas, J. et al. Germline copy number variation of genes involved in chromatin remodelling in families suggestive of Li–Fraumeni syndrome with brain tumours. Eur. J. Hum. Genet. 21, 1369–1376 (2013).
Alhazzazi, T. Y. et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer 117, 1670–1678 (2011).
Jeong, S. M. et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23, 450–463 (2013). This paper reveals the requirement of the mitochondrial sirtuin SIRT4 for inhibiting glutamine metabolism and ensuring proper DNA repair upon DNA damage.
Jeong, S. M., Lee, A., Lee, J. & Haigis, M. C. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J. Biol. Chem. 289, 4135–4144 (2014).
Garber, M. E. et al. Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl Acad. Sci. USA 98, 13784–13789 (2001).
Choi, Y. L. et al. A genomic analysis of adult T-cell leukemia. Oncogene 26, 1245–1255 (2007).
Wang, Q. et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med. Oncol. 29, 77–83 (2012).
Blaveri, E. et al. Bladder cancer outcome and subtype classification by gene expression. Clin. Cancer Res. 11, 4044–4055 (2005).
Haigis, M. C. et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 126, 941–954 (2006).
Csibi, A. et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153, 840–854 (2013). This study demonstrates the relationship between high levels of mTORC1 signalling, which is a hallmark of cancer, and reduced levels of SIRT4.
Lu, W., Zuo, Y., Feng, Y. & Zhang, M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol. 35, 10699–10705 (2014).
Michishita, E. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 (2008). This paper demonstrates that SIRT6 is localized at telomeric chromatin and is required for the stable association of WRN and the maintenance of telomeric structures.
Kawahara, T. L. et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136, 62–74 (2009).
Michishita, E. et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8, 2664–2666 (2009).
Kugel, S. & Mostoslavsky, R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 39, 72–81 (2014).
Mostoslavsky, R. et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 (2006). This paper shows that SIRT6 is necessary for genomic stability as it regulates the BER pathway. SIRT6 absence in mice leads to progeria phenotypes and premature death.
Zhong, L. et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 (2010).
Yang, B., Zwaans, B. M., Eckersdorff, M. & Lombard, D. B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663 (2009).
Sebastian, C. et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151, 1185–1199 (2012). This study reports that SIRT6 functions as a tumour suppressor by regulating aerobic glycolysis and glutamine metabolism in cancer cells.
Min, L. et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat. Cell Biol. 14, 1203–1211 (2012).
Marquardt, J. U. et al. Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology 58, 1054–1064 (2013).
Khongkow, M. et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis 34, 1476–1486 (2013).
Bauer, I. et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J. Biol. Chem. 287, 40924–40937 (2012).
Chen, S. et al. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell 52, 303–313 (2013).
Grob, A. et al. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 122, 489–498 (2009).
Barber, M. F. et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114–118 (2012). This report shows that SIRT7 regulates H3K18 acetylation at gene promoters, represses transcription and promotes oncogenesis.
Warburg, O. On respiratory impairment in cancer cells. Science 124, 269–270 (1956).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Zwaans, B. M. & Lombard, D. B. Interplay between sirtuins, MYC and hypoxia-inducible factor in cancer-associated metabolic reprogramming. Dis. Model. Mech. 7, 1023–1032 (2014).
Majmundar, A. J., Wong, W. J. & Simon, M. C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294–309 (2010).
Qiu, X., Brown, K., Hirschey, M. D., Verdin, E. & Chen, D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell. Metab. 12, 662–667 (2010).
Tao, R. et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 40, 893–904 (2011).
Bell, E. L., Emerling, B. M., Ricoult, S. J. & Guarente, L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene 30, 2986–2996 (2011). This study demonstrates that SIRT3 functions as a tumour suppressor by reducing ROS production and destabilizing HIF1α.
Richardson, D. R., Kalinowski, D. S., Lau, S., Jansson, P. J. & Lovejoy, D. B. Cancer cell iron metabolism and the development of potent iron chelators as anti-tumour agents. Biochim. Biophys. Acta 1790, 702–717 (2009).
Yang, H. et al. SIRT3-dependent GOT2 acetylation status affects the malate-aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J. 34, 1110–1125 (2015).
Fan, J. et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol. Cell 53, 534–548 (2014).
Ozden, O. et al. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radic. Biol. Med. 76, 163–172 (2014).
Eilers, M. & Eisenman, R. N. Myc's broad reach. Genes Dev. 22, 2755–2766 (2008).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Mao, B. et al. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int. J. Biochem. Cell Biol. 43, 1573–1581 (2011).
Marshall, G. M. et al. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet. 7, e1002135 (2011).
Yuan, J., Minter-Dykhouse, K. & Lou, Z. A. c-Myc–SIRT1 feedback loop regulates cell growth and transformation. J. Cell Biol. 185, 203–211 (2009).
Liu, P. Y. et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 20, 503–514 (2013).
Lin, Z. et al. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 5, 1639–1649 (2013).
Jochemsen, A. G. & Shiloh, Y. USP10: friend and foe. Cell 140, 308–310 (2010).
Shin, J. et al. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 5, 654–665 (2013).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Hoeijmakers, J. H. Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 (2001).
Xu, Z. et al. SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle 14, 269–276 (2015).
Fan, W. & Luo, J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol. Cell 39, 247–258 (2010).
Ming, M. et al. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc. Natl Acad. Sci. USA 107, 22623–22628 (2010). References 113 and 114 show that SIRT1 plays an important part in the NER pathway by activating the XPA and XPC repair factors, through deacetylation and induced gene expression, respectively.
Fang, E. F. et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell 157, 882–896 (2014).
Scheibye-Knudsen, M. et al. A high-fat diet and NAD+ activate Sirt1 to rescue premature aging in Cockayne syndrome. Cell. Metab. 20, 840–855 (2014).
Wang, R. H. et al. SIRT1 deacetylates TopBP1 and modulates intra-S-phase checkpoint and DNA replication origin firing. Int. J. Biol. Sci. 10, 1193–1202 (2014).
Chapman, J. R., Taylor, M. R. & Boulton, S. J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012).
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
San Filippo, J., Sung, P. & Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008).
Palacios, J. A. et al. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J. Cell Biol. 191, 1299–1313 (2010).
Yuan, Z., Zhang, X., Sengupta, N., Lane, W. S. & Seto, E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell 27, 149–162 (2007).
Li, K. et al. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J. Biol. Chem. 283, 7590–7598 (2008).
Toiber, D. et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 51, 454–468 (2013).
Mao, Z. et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446 (2011). This article demonstrates that SIRT6 is recruited after oxidative stress to the sites of DNA DSBs and stimulates DNA repair by mono-ADP-ribosylating PARP1 and stimulating its poly-ADP-ribosylase activity.
Kaidi, A., Weinert, B. T., Choudhary, C. & Jackson, S. P. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329, 1348–1353 (2010). This study shows that SIRT6 plays an important part in enhancing DSB repair by deacetylating CtIP and promoting DNA resection.
Cohen, H. Y. et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–392 (2004).
Jeong, J. et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp. Mol. Med. 39, 8–13 (2007).
McCord, R. A. et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging 1, 109–121 (2009).
Dioum, E. M. et al. Regulation of hypoxia-inducible factor 2α signaling by the stress-responsive deacetylase sirtuin 1. Science 324, 1289–1293 (2009).
Lim, J. H. et al. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1α. Mol. Cell 38, 864–878 (2010).
Laemmle, A. et al. Inhibition of SIRT1 impairs the accumulation and transcriptional activity of HIF-1α protein under hypoxic conditions. PLoS ONE 7, e33433 (2012).
Guarani, V. et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature 473, 234–238 (2011).
Xie, M., Liu, M. & He, C. S. SIRT1 regulates endothelial Notch signaling in lung cancer. PLoS ONE 7, e45331 (2012).
Hubbi, M. E., Hu, H., Kshitiz, Gilkes, D. M. & Semenza, G. L. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J. Biol. Chem. 288, 20768–20775 (2013).
Suzuki, K. et al. SRT1720, a SIRT1 activator, promotes tumor cell migration, and lung metastasis of breast cancer in mice. Oncol. Rep. 27, 1726–1732 (2012).
Byles, V. et al. SIRT1 induces EMT by cooperating with EMT transcription factors and enhances prostate cancer cell migration and metastasis. Oncogene 31, 4619–4629 (2012).
Malik, S. et al. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci. Rep. 5, 9841 (2015).
Van Gool, F. et al. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat. Med. 15, 206–210 (2009).
Jiang, H. et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 (2013).
Xiao, C. et al. Progression of chronic liver inflammation and fibrosis driven by activation of c-JUN signaling in Sirt6 mutant mice. J. Biol. Chem. 287, 41903–41913 (2012).
Yeung, F. et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380 (2004).
Kojima, K. et al. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem. Biophys. Res. Commun. 373, 423–428 (2008).
Wang, J. et al. Sirtinol, a class III HDAC inhibitor, induces apoptotic and autophagic cell death in MCF-7 human breast cancer cells. Int. J. Oncol. 41, 1101–1109 (2012).
Lain, S. et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 13, 454–463 (2008).
He, X. et al. SIRT2 activity is required for the survival of C6 glioma cells. Biochem. Biophys. Res. Commun. 417, 468–472 (2012).
Hoffmann, G., Breitenbucher, F., Schuler, M. & Ehrenhofer-Murray, A. E. A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J. Biol. Chem. 289, 5208–5216 (2014).
Banerjee, S., Kaye, S. B. & Ashworth, A. Making the best of PARP inhibitors in ovarian cancer. Nat. Rev. Clin. Oncol. 7, 508–519 (2010).
Gottlieb, S. & Esposito, R. E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56, 771–776 (1989).
Bryk, M. et al. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11, 255–269 (1997).
Guarente, L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14, 1021–1026 (2000).
Braunstein, M., Sobel, R. E., Allis, C. D., Turner, B. M. & Broach, J. R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16, 4349–4356 (1996).
Kennedy, B. K., Austriaco, N. R. Jr., Zhang, J. & Guarente, L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80, 485–496 (1995).
Kennedy, B. K. et al. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89, 381–391 (1997).
Sinclair, D. A. & Guarente, L. Extrachromosomal rDNA circles — a cause of aging in yeast. Cell 91, 1033–1042 (1997).
Kaeberlein, M., McVey, M. & Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 (1999).
Houtkooper, R. H., Pirinen, E. & Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 13, 225–238 (2012).
North, B. J., Marshall, B. L., Borra, M. T., Denu, J. M. & Verdin, E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444 (2003).
Li, W. et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating α-tubulin. J. Neurosci. 27, 2606–2616 (2007).
Jing, E., Gesta, S. & Kahn, C. R. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell. Metab. 6, 105–114 (2007).
Wang, Y. P. et al. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J. 33, 1304–1320 (2014).
Xu, Y. et al. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 74, 3630–3642 (2014).
Bell, E. L. & Guarente, L. The SirT3 divining rod points to oxidative stress. Mol. Cell 42, 561–568 (2011).
Hallows, W. C. et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell 41, 139–149 (2011).
Lombard, D. B. et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807–8814 (2007).
Laurent, G. et al. SIRT4 represses peroxisome proliferator-activated receptor α activity to suppress hepatic fat oxidation. Mol. Cell. Biol. 33, 4552–4561 (2013).
Laurent, G. et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol. Cell 50, 686–698 (2013).
Mathias, R. A. et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 159, 1615–1625 (2014).
Peng, C. et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, M111.012658 (2011).
Park, J. et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 (2013).
Rardin, M. J. et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell. Metab. 18, 920–933 (2013).
Tan, M. et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell. Metab. 19, 605–617 (2014).
Nishida, Y. et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell 59, 321–332 (2015).
Kanfi, Y. et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 (2012).
Ryu, D. et al. A SIRT7-dependent acetylation switch of GABPβ1 controls mitochondrial function. Cell. Metab. 20, 856–869 (2014).
Yoshizawa, T. et al. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin–proteasome pathway. Cell. Metab. 19, 712–721 (2014).
Vakhrusheva, O. et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703–710 (2008).
Acknowledgements
The authors thank E. Williams for comments on the manuscript. They apologize to colleagues whose original work could not be cited owing to space limitations. L.G. is supported by the US National Institutes of Health and the Glenn Foundation for Medical Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
L.G. consults for GlaxoSmithKline, Chronos Therapeutics and Segterra, and he is a founder of Elysium Health. A.C. declares no competing interests.
Related links
DATABASES
Glossary
- Glycolysis
-
The conversion of glucose into pyruvate in a series of enzymatic steps, with the concurrent generation of ATP.
- Glutaminolysis
-
A series of enzymatic reactions in which the amino acid glutamine is converted into glutamate, which can be further metabolized to α-ketoglutarate.
- Calorie restriction
-
A dietary regimen in which all essential nutrients are supplied but calories are reduced by 20–40% compared to ad libitum feeding.
- Oxidative metabolism
-
The oxidation of carbohydrates to CO2 in mitochondria to maximize the production of ATP.
- Thymocytes
-
Haematopoietic progenitor cells that reside in the thymus and differentiate into mature T lymphocytes.
- Enterocyte
-
A type of epithelial cell that resides in the intestine and absorbs nutrients.
- Pentose phosphate shunt
-
An anabolic pathway that converts glucose into pentoses (five-carbon sugars used in nucleotide synthesis), with the concurrent generation of NADPH.
- E boxes
-
DNA sequences found in promoters and recognized by transcription factors that contain the basic helix–loop–helix motif.
- Pyrimidine dimers
-
Covalent linkages of thymine or cytosine bases in DNA that are induced by ultraviolet light.
- Interstrand crosslinks
-
Covalent linkages of complementary strands of DNA that are induced by various exogenous agents (for example, cisplatin) or endogenous agents (for example, aldehydes).
- Telomeres
-
Repetitive nucleotide sequences at the ends of chromosomes that protect them from degradation and fusion with neighbouring chromosomes.
- Centromeres
-
Regions of chromosomes at which two sister chromatids attach to the mitotic spindle for proper segregation during mitosis.
- γH2AX
-
The phosphorylated form of the histone variant H2AX, which marks double-stranded DNA breaks.
Rights and permissions
About this article
Cite this article
Chalkiadaki, A., Guarente, L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer 15, 608–624 (2015). https://doi.org/10.1038/nrc3985
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3985
This article is cited by
-
Acylspermidines are conserved mitochondrial sirtuin-dependent metabolites
Nature Chemical Biology (2024)
-
Dysregulation of histone deacetylases in ocular diseases
Archives of Pharmacal Research (2024)
-
Role of sirtuins in attenuating plaque vulnerability in atherosclerosis
Molecular and Cellular Biochemistry (2024)
-
Ochratoxin A induces abnormal tryptophan metabolism in the intestine and liver to activate AMPK signaling pathway
Journal of Animal Science and Biotechnology (2023)
-
Sirt3 restricts tumor initiation via promoting LONP1 deacetylation and K63 ubiquitination
Journal of Translational Medicine (2023)