Key Points
-
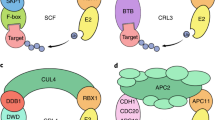
COP1 is a well-conserved E3 ubiquitin ligase that is involved in the ubiquitylation of various protein substrates to trigger their proteasomal degradation.
-
Several putative targets of mouse and human COP1 have been identified, including COP1 itself, p53, JUN and ETS variant (ETV) family members, indicating that depending on context it may act as an oncogene or as a tumour suppressor.
-
Both overexpression and loss of function of COP1 have been described in a variety of human tumours. In mice, the complete loss of COP1 function results in embryonic lethality, whereas studies of a partial loss of COP1 function have provided the first in vivo evidence for its tumour suppressor function.
-
Most identified COP1 targets are transcription factors that are implicated in cancer, although other targets imply that COP1 might also function in lipid and glucose metabolism. The main cellular functions of COP1 are probably mediated through the ubiquitylation and degradation of its substrates.
-
Unlike previously described E3 ligases, such as FBW7 and ITCH, COP1 can target unphosphorylated JUN for degradation. Thus, distinct pools of JUN may be targeted for degradation by different E3 ligases.
-
Given its role in the regulation of JUN and of the ETV family members, which are all known prostate cancer oncoproteins, COP1 might have a particularly important role in prostate cancer.
-
Despite early biochemical evidence that COP1 targets p53, recent in vivo data suggest that this interaction is unlikely to have an important role or to even occur in vivo.
-
The absence of confirmatory evidence for a role for COP1 in the regulation of p53 stability and activity argues against the use of COP1-inhibitory drugs for cancer therapy.
Abstract
COP1 is an E3 ubiquitin ligase that is involved in the ubiquitylation of various protein substrates to trigger their proteasomal degradation. Although originally identified in a light signalling pathway in plants, biochemical studies have identified putative targets of mammalian COP1 with relevant roles in tumorigenesis, including the oncoproteins JUN and ETV family members, as well as the p53 tumour suppressor. Recent genetic studies have shown that COP1 deficiency leads to spontaneous tumour formation in mice, and have identified mutations in COP1 and its substrates in various human cancers. These findings add to our growing appreciation of the roles for E3 ligases in cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wang, H., Kang, D., Deng, X. W. & Wei, N. Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Curr. Biol. 9, 711–714 (1999).
Deng, X. W., Caspar, T. & Quail, P. H. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5, 1172–1182 (1991). This paper identifies COP1 for the first time as a genetic locus in A. thaliana and provides a link between COP1 and light signalling.
Scheffner, M., Nuber, U. & Huibregtse, J. M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373, 81–83 (1995).
Chau, V. et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 (1989).
Thrower, J. S., Hoffman, L., Rechsteiner, M. & Pickart, C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 (2000).
Hicke, L., Schubert, H. L. & Hill, C. P. Ubiquitin-binding domains. Nature Rev. Mol. Cell Biol. 6, 610–621 (2005).
Eddins, M. J., Carlile, C. M., Gomez, K. M., Pickart, C. M. & Wolberger, C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nature Struct. Mol. Biol. 13, 915–920 (2006).
Rodrigo-Brenni, M. C., Foster, S. A. & Morgan, D. O. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell 39, 548–559 (2010).
Wickliffe, K. E., Lorenz, S., Wemmer, D. E., Kuriyan, J. & Rape, M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 (2011).
Zheng, N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000).
Hoeller, D. & Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 (2009).
Nakayama, K. I. & Nakayama, K. Ubiquitin ligases: cell-cycle control and cancer. Nature Rev. Cancer 6, 369–381 (2006).
Lipkowitz, S. & Weissman, A. M. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nature Rev. Cancer 11, 629–643 (2011).
Welcker, M. & Clurman, B. E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature Rev. Cancer 8, 83–93 (2008).
Frescas, D. & Pagano, M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nature Rev. Cancer 8, 438–449 (2008).
Savio, M. G et al. COP1D, an alternatively spliced constitutive photomorphogenic-1 (COP1) product, stabilizes UV stress-induced c-Jun through inhibition of full-length COP1. Oncogene 27, 2401–2411 (2008).
Wertz, I. E. et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303, 1371–1374 (2004). Provides further evidence that COP1 targets JUN and shows that COP1 acts as a bridge between JUN, DET1 and the large E3 ligase complex that contains CUL4A.
Dornan, D. et al. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 64, 7226–7230 (2004).
Lee, Y. H. et al. Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer Res. 70, 8264–8269 (2010).
Su, C. H. et al. 14-3-3sigma exerts tumor-suppressor activity mediated by regulation of COP1 stability. Cancer Res. 71, 884–894 (2011).
Migliorini, D. et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J. Clin. Invest. 121, 1329–1343 (2011). The characterization of hypomorphic Cop1 mouse phenotypes provided evidence that COP1 targets JUN but not p53 for degradation in vivo and therefore acts as a tumour suppressor.
Vitari, A. C. et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474, 403–406 (2011). An in vivo demonstration that COP1 targets members of the ETV family of transcription factors and further evidence for the importance of COP1 in the aetiology of prostate cancer.
Baxter, L. L., Hou, L., Loftus, S. K. & Pavan, W. J. Spotlight on spotted mice: a review of white spotting mouse mutants and associated human pigmentation disorders. Pigment Cell Res. 17, 215–224 (2004).
Wei, W. & Kaelin, W. G., Jr. Good COP1 or bad COP1? In vivo veritas. J. Clin. Invest. 121, 1263–1265 (2011).
Dornan, D. et al. ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 313, 1122–1126 (2006).
Su, C. H. et al. Nuclear export regulation of COP1 by 14-3-3sigma in response to DNA damage. Mol. Cancer 9, 243 (2010).
Yi, C., Wang, H., Wei, N. & Deng, X. W. An initial biochemical and cell biological characterization of the mammalian homologue of a central plant developmental switch, COP1. BMC Cell Biol. 3, 30 (2002). The first demonstration of a structural and potentially functional conservation of COP1 between A. thaliana , mouse and human. It provided the first evidence that mammalian COP1, similarly to its plant homologue, may have a role in ubiquitylation.
Fu, H., Reis, N., Lee, Y., Glickman, M. H. & Vierstra, R. D. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20, 7096–7107 (2001).
Wei, N. & Deng, X. W. Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 15, 98–103 (1999).
Kato, J. Y. & Yoneda-Kato, N. Mammalian COP9 signalosome. Genes Cells 14, 1209–1225 (2009).
Yoneda-Kato, N., Tomoda, K., Umehara, M., Arata, Y. & Kato, J. Y. Myeloid leukemia factor 1 regulates p53 by suppressing COP1 via COP9 signalosome subunit 3. EMBO J. 24, 1739–1749 (2005).
Choi, H. H. et al. COP9 signalosome subunit 6 stabilizes COP1, which functions as an E3 ubiquitin ligase for 14-3-3sigma. Oncogene 30, 4791–4801 (2011).
Richardson, K. S. & Zundel, W. The emerging role of the COP9 signalosome in cancer. Mol. Cancer Res. 3, 645–653 (2005).
Lee, M. H., Zhao, R., Phan, L. & Yeung, S. C. Roles of COP9 signalosome in cancer. Cell Cycle 10, 3057–3066 (2011).
Adler, A. S. et al. CSN5 isopeptidase activity links COP9 signalosome activation to breast cancer progression. Cancer Res. 68, 506–515 (2008).
Mori, M., Yoneda-Kato, N., Yoshida, A. & Kato, J. Y. Stable form of JAB1 enhances proliferation and maintenance of hematopoietic progenitors. J. Biol. Chem. 283, 29011–29021 (2008).
Zhao, R. et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J. Clin. Invest. 121, 851–865 (2011).
Bianchi, E. et al. Characterization of human constitutive photomorphogenesis protein 1, a RING finger ubiquitin ligase that interacts with Jun transcription factors and modulates their transcriptional activity. J. Biol. Chem. 278, 19682–19690 (2003). Is supported by reference 17 and is the first biochemical demonstration that COP1 targets JUN. The results indicate that human COP1 is a regulator of AP1-dependent transcription that shares important properties of A. thaliana COP1 in the control of gene expression.
Dornan, D. et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429, 86–92 (2004). Biochemical evidence that p53 is a target of COP1.
Derijard, B. et al. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76, 1025–1037 (1994).
Kyriakis, J. M. et al. The stress-activated protein kinase subfamily of c-Jun kinases. Nature 369, 156–160 (1994).
Arias, J. et al. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370, 226–229 (1994).
Bannister, A. J., Oehler, T., Wilhelm, D., Angel, P. & Kouzarides, T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene 11, 2509–2514 (1995).
Karin, M. The regulation of AP-1 activity by mitogen-activated protein kinases. J. Biol. Chem. 270, 16483–16486 (1995).
Zhong, S. P., Ma, W. Y., Quealy, J. A., Zhang, Y. & Dong, Z. Organ-specific distribution of AP-1 in AP-1 luciferase transgenic mice during the maturation process. Am. J. Physiol. Regul. Integr Comp. Physiol. 280, R376–R381 (2001).
Huang, C. et al. Inhibition of ultraviolet B-induced activator protein-1 (AP-1) activity by aspirin in AP-1-luciferase transgenic mice. J. Biol. Chem. 272, 26325–26331 (1997).
Nateri, A. S., Riera-Sans, L., Da Costa, C. & Behrens, A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303, 1374–1378 (2004).
Wei, W., Jin, J., Schlisio, S., Harper, J. W. & Kaelin, W. G., Jr. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 (2005).
Gao, M. et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306, 271–275 (2004). References 47–49 describe the complexity of JUN regulation by several different E3 ligases.
Fang, D. & Kerppola, T. K. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc. Natl Acad. Sci. USA 101, 14782–14787 (2004).
Yi, C. et al. Major vault protein, in concert with constitutively photomorphogenic 1, negatively regulates c-Jun-mediated activator protein 1 transcription in mammalian cells. Cancer Res. 65, 5835–5840 (2005).
Fuchs, S. Y., Dolan, L., Davis, R. J. & Ronai, Z. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene 13, 1531–1535 (1996).
Musti, A. M., Treier, M. & Bohmann, D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 275, 400–402 (1997).
Davis, R. J. Signal transduction by the JNK group of MAP kinases. Cell 103, 239–252 (2000).
Edwards, J., Krishna, N. S., Mukherjee, R. & Bartlett, J. M. The role of c-Jun and c-Fos expression in androgen-independent prostate cancer. J. Pathol. 204, 153–158 (2004).
Chen, S. Y. et al. c-Jun enhancement of androgen receptor transactivation is associated with prostate cancer cell proliferation. Oncogene 25, 7212–7223 (2006).
Ouyang, X. et al. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 68, 2132–2144 (2008).
Barcellos-Hoff, M. H., Park, C. & Wright, E. G. Radiation and the microenvironment - tumorigenesis and therapy. Nature Rev. Cancer 5, 867–875 (2005).
Chi, P. et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature 467, 849–853 (2010).
Jane-Valbuena, J. et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 70, 2075–2084 (2010).
Kumar-Sinha, C., Tomlins, S. A. & Chinnaiyan, A. M. Recurrent gene fusions in prostate cancer. Nature Rev. Cancer 8, 497–511 (2008).
Baert, J. L. et al. The E3 ubiquitin ligase complex component COP1 regulates PEA3 group member stability and transcriptional activity. Oncogene 29, 1810–1820 (2010). The first biochemical demonstration that COP1 regulates ETV1 and its family members.
Tomlins, S. A. et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448, 595–599 (2007).
Schreiber, M. et al. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 13, 607–619 (1999).
Migliorini, D. et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol. Cell Biol. 22, 5527–5538 (2002).
Francoz, S et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc. Natl Acad. Sci. USA 103, 3232–3237 (2006).
Marine, J. C. et al. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 13, 927–934 (2006).
Fu, X. et al. RFWD3-Mdm2 ubiquitin ligase complex positively regulates p53 stability in response to DNA damage. Proc. Natl Acad. Sci. USA 107, 4579–4584 (2010).
Amson, R et al. Reciprocal repression between P53 and TCTP. Nature Med. 18, 91–99 (2011).
Mendrysa, S. M. et al. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol. Cell Biol. 23, 462–472 (2003).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Horn, H. F. & Vousden, K. H. Coping with stress: multiple ways to activate p53. Oncogene 26, 1306–1316 (2007).
Li, D. Q et al. E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc. Natl Acad. Sci. USA 106, 17493–17498 (2009).
Liu, Y. et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature 456, 269–273 (2008). This reference describes for the first time a putative role for COP1 in glucose metabolism.
Koo, S. H. et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437, 1109–1111 (2005).
Kato, S., Ding, J., Pisck, E., Jhala, U. S. & Du, K. COP1 functions as a FoxO1 ubiquitin E3 ligase to regulate FoxO1-mediated gene expression. J. Biol. Chem. 283, 35464–35473 (2008).
Zhang, Y., Gan, B., Liu, D. & Paik, J. H. FoxO family members in cancer. Cancer Biol. Ther. 12, 253–259 (2011).
Sykes, S. H. H. et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell 146, 697–708 (2011).
Qi, L. et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 312, 1763–1766 (2006). This reference describes for the first time a putative role for COP1 in lipid metabolism.
Beckers, A. et al. Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 67, 8180–8187 (2007).
Weinstein, I. B. Cancer. Addiction to oncogenes- the Achilles heal of cancer. Science 297, 63–64 (2002).
Sullivan, J. A., Shirasu, K. & Deng, X.W. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nature Rev. Genet. 4, 948–958 (2003).
Osterlund, M. T., Hardtke, C. S. Wei, N. & Deng, X. W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 (2000).
Saijo, Y. et al. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642–2647 (2003).
Yi, C. & Deng, X. W. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625 (2005).
Wang, X. et al. Regulation of COP1 nuclear localization by the COP9 signalosome via direct interaction with CSN1. Plant J. 58, 655–667 (2009).
Schwechheimer, C. The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim. Biophys. Acta 1695, 45–54 (2004).
Schwechheimer, C. & Isono, E. The COP9 signalosome and its role in plant development. Eur. J. Cell Biol. 89, 157–162 (2010).
Saha, A. & Deshaies, R. J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 (2008).
Dohmann, E. M. et al. The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135, 2013–2022 (2008).
Hannss, R. & Dubiel, W. COP9 signalosome function in the DDR. FEBS Lett. 585, 2845–2852 (2011).
Suzuki, G., Yanagawa, Y., Kwok, S. F., Matsui, M. & Deng, X. W. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16, 554–559 (2002).
Yanagawa, Y. et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18, 2172–2181 (2004).
Jackson, S. & Xiong, Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34, 562–570 (2009).
Seo, H. S. et al. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995–999 (2003). The first evidence that COP1 acts as an intrinsic E3 ligase and the identification of COP1 itself and the MYB transcription activator LAF1 as the first targets of COP1.
Chang, L. et al. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell 124, 601–613 (2006).
Acknowledgements
The author thanks M. Skipper for help in preparing this manuscript. This research was partly supported by VIB, the Belgian Federation for Cancer and AICR.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Glossary
- 26S proteasome
-
The ATP-dependent proteolytic complex that is responsible for ubiquitin-dependent protein degradation.
- RING domain
-
A protein structural domain of zinc finger proteins, which contains a (Cys)3-His-(Cys)4 amino acid motif that binds two zinc cations. It contains 40 to 60 amino acids. Many proteins containing a RING domain are involved in the ubiquitylation pathway.
- WD40 repeats
-
Also known as the WD repeats and β-transducin repeats. Short structural motifs of approximately 40 amino acids, with tryptophan and aspartic acid often found at their ends; several such repeats combine to form the WD domain.
- Hydrops fetalis
-
A condition in which abnormal amounts of fluid accumulate in two or more body areas of a fetus or newborn.
- Pleiotropic
-
A phenomenon whereby one gene influences multiple phenotypic traits.
- Histiocytic sarcoma
-
A tumour derived from histiocytes, cells that are part of the mononuclear phagocyte system.
- Testicular teratoma
-
A type of gonadal tumour derived from germ cells that occurs in the testes.
- Degrons
-
Sequences of amino acids in a protein that is necessary and sufficient to confer its degradation by the ubiquitin–proteasome system.
Rights and permissions
About this article
Cite this article
Marine, JC. Spotlight on the role of COP1 in tumorigenesis. Nat Rev Cancer 12, 455–464 (2012). https://doi.org/10.1038/nrc3271
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3271
This article is cited by
-
E26 transformation-specific transcription variant 5 in development and cancer: modification, regulation and function
Journal of Biomedical Science (2023)
-
CUL4B-DDB1-COP1-mediated UTX downregulation promotes colorectal cancer progression
Experimental Hematology & Oncology (2023)
-
Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination
Communications Biology (2023)
-
E3 ubiquitin ligase COP1-mediated CEBPB ubiquitination regulates the inflammatory response of macrophages in sepsis-induced myocardial injury
Mammalian Genome (2023)
-
The E3 ligase COP1 promotes ERα signaling and suppresses EMT in breast cancer
Oncogene (2022)