Key Points
-
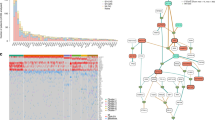
The initiation of myeloma is mediated by the interaction of environmental factors and inherited genetic events that, when combined with the normal physiological processes that are necessary to generate antibody diversity, result in genetic changes that lead to the immortalization of a myeloma-propagating cell. Chief among these events are chromosomal translocations and hyperdiploidy.
-
Based on their distribution in most clonal cells, chromosomal translocations that are generated by aberrant class switch recombination have been suggested to be initiating events occurring early in the disease process. As a result of these translocations, oncogenes can be placed under the control of the strong enhancers of the immunoglobulin gene loci, leading to their increased expression.
-
The interaction of normal plasma cells with their supportive microenvironment is crucial for plasma cell longevity. A characteristic feature of myeloma cells is the requirement for an intimate relationship with the bone marrow microenvironment, where plasma cells are nurtured in specialized niches that facilitate the growth of the myeloma clone. Derangement of these interactions is important in the immortalization of a myeloma-propagating cell.
-
The basic premise underlying the progression of myeloma is that multiple mutations in different pathways collaborate to deregulate the intrinsic biology of the plasma cell, changing it in ways that lead to the features of myeloma. The immortalized cell then acquires additional genetic hits over time, thus leading to the clinically recognized features of myeloma and eventually to the development of treatment resistance and an ability to grow in the peripheral blood as a leukaemic phase.
-
It is becoming increasingly clear that the molecular events acquired during myeloma progression are not acquired in a linear fashion but instead through branching, nonlinear pathways, similar to the mechanism suggested by Darwin to explain the evolution of species. Therefore, a further level of the genetic complexity of myeloma is based on intraclonal heterogeneity at the level of a myeloma-propagating cell.
-
The genetic lesions that lead to myeloma are best considered within the categories of inherited variation, translocations, copy number abnormalities, mutations, and methylation and miRNA abnormalities. The net biological impact of such events is to modify the behaviour of the myeloma-propagating cell, resulting in the key molecular hallmarks of myeloma.
-
The treatments that are used for myeloma include steroids, alkylating agents, the immunomodulatory drugs thalidomide and lenalidomide, proteasome inhibitors and autologous stem cell transplantation. Current therapeutic aims are to induce and maintain long-term remission. Future approaches based on personalized medicine strategies will involve targeted therapies combined with molecular-diagnostic strategies.
-
The presence of intraclonal heterogeneity has important effects on the clinical application of both standard and targeted treatment strategies but provides a model system within which to assess their optimum use.
Abstract
Based on the clinical features of myeloma and related malignancies of plasma cells, it has been possible to generate a model system of myeloma progression from a normal plasma cell through smouldering myeloma to myeloma and then plasma cell leukaemia. Using this model system we can study at which points the genetic alterations identified through whole-tumour molecular analyses function in the initiation and progression of myeloma. Further genetic complexity, such as intraclonal heterogeneity, and insights into the molecular evolution and intraclonal dynamics in this model system are crucial to our understandings of tumour progression, treatment resistance and the use of currently available and future treatments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Greipp, P. R. et al. International staging system for multiple myeloma. J. Clin. Oncol. 23, 3412–3420 (2005).
Kyle, R. A. et al. Review of 1027 patients with newly diagnosed multiple myeloma. Mayo Clin. Proc. 78, 21–33 (2003).
Kyle, R. A. et al. Clinical course and prognosis of smoldering (asymptomatic) multiple myeloma. N. Engl. J. Med. 356, 2582–2590 (2007).
Akiyama, M. et al. Cytokines modulate telomerase activity in a human multiple myeloma cell line. Cancer Res. 62, 3876–3882 (2002).
Chauhan, D. et al. Blockade of ubiquitin-conjugating enzyme CDC34 enhances anti-myeloma activity of Bortezomib/Proteasome inhibitor PS-341. Oncogene 23, 3597–3602 (2004).
Damiano, J. S., Cress, A. E., Hazlehurst, L. A., Shtil, A. A. & Dalton, W. S. Cell adhesion mediated drug resistance (CAM-DR): role of integrins and resistance to apoptosis in human myeloma cell lines. Blood 93, 1658–1667 (1999).
Hideshima, T. et al. Perifosine, an oral bioactive novel alkylphospholipid, inhibits Akt and induces in vitro and in vivo cytotoxicity in human multiple myeloma cells. Blood 107, 4053–4062 (2006).
Mitsiades, C. S., Mitsiades, N. S., Munshi, N. C., Richardson, P. G. & Anderson, K. C. The role of the bone microenvironment in the pathophysiology and therapeutic management of multiple myeloma: interplay of growth factors, their receptors and stromal interactions. Eur. J. Cancer 42, 1564–1573 (2006).
Palumbo, A. & Anderson, K. Multiple myeloma. N. Engl. J. Med. 364, 1046–1060 (2011).
Chapman, M. A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011). The first report of a myeloma genome, which demonstrated the complex and varied abnormalities that are seen in myeloma.
Gonzalez, D. et al. Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma. Blood 110, 3112–3121 (2007). A comprehensive review of normal antibody generation and plasma cell development, detailing the changes that are seen in MGUS and myeloma. The importance of SHM and CSR is highlighted.
Bergsagel, P. L. & Kuehl, W. M. Critical roles for immunoglobulin translocations and cyclin D dysregulation in multiple myeloma. Immunol. Rev. 194, 96–104 (2003). A review explaining the biological significance of chromosome 14 translocations in myeloma.
Hakim, O. et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 7 Feb 2012 (doi:10.1038/nature10909).
Reimold, A. M. et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412, 300–307 (2001).
Iwakoshi, N. N. et al. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nature Immunol. 4, 321–329 (2003).
Shaffer, A. L. et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93 (2004).
Davenport, E. L., Morgan, G. J. & Davies, F. E. Untangling the unfolded protein response. Cell Cycle 7, 865–869 (2008).
Carrasco, D. R. et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 11, 349–360 (2007).
Bagratuni, T. et al. XBP1s levels are implicated in the biology and outcome of myeloma mediating different clinical outcomes to thalidomide-based treatments. Blood 116, 250–253 (2010).
Oracki, S. A., Walker, J. A., Hibbs, M. L., Corcoran, L. M. & Tarlinton, D. M. Plasma cell development and survival. Immunol. Rev. 237, 140–159 (2010).
Radbruch, A. et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nature Rev. Immunol. 6, 741–750 (2006).
Chiecchio, L. et al. Timing of acquisition of deletion 13 in plasma cell dyscrasias is dependent on genetic context. Haematologica 94, 1708–1713 (2009).
Carrasco, D. R. et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell 9, 313–325 (2006).
Walker, B. A. et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood 116, e56–e65 (2010).
Lopez-Corral, L. et al. The progression from MGUS to smoldering myeloma and eventually to multiple myeloma involves a clonal expansion of genetically abnormal plasma cells. Clin. Cancer Res. 17, 1692–1700 (2011).
Cremer, F. W. et al. High incidence and intraclonal heterogeneity of chromosome 11 aberrations in patients with newly diagnosed multiple myeloma detected by multiprobe interphase FISH. Cancer Genet. Cytogenet. 161, 116–124 (2005).
Walker, B. A. et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms and genes important in the pathogenesis of multiple myeloma. Blood 108, 1733–1743 (2006).
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011). An important paper discussing the role of intraclonal variation in tumour biology.
Sahota, S. S., Leo, R., Hamblin, T. J. & Stevenson, F. K. Ig VH gene mutational patterns indicate different tumor cell status in human myeloma and monoclonal gammopathy of undetermined significance. Blood 87, 746–755 (1996).
Ross, F. M. et al. Age has a profound effect on the incidence and significance of chromosome abnormalities in myeloma. Leukemia 19, 1634–1642 (2005).
Akiyama, M. et al. Effects of oligonucleotide N3′→P5′ thio-phosphoramidate (GRN163) targeting telomerase RNA in human multiple myeloma cells. Cancer Res. 63, 6187–6194 (2003).
Shammas, M. A. et al. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clin. Cancer Res. 10, 770–776 (2004).
Brennan, S. K. et al. Telomerase inhib ition targets clonogenic multiple myeloma cells through telomere length-dependent and independent mechanisms. PLoS ONE 5, e12487 (2010).
McKenna, R. W. et al. Plasma cell neoplasms. In World Health Organization Classification of Tumors of Hematopoietic and Lymphoid Tissues (eds Swerdlow, S. H. et al.) 200–213 (IARC Press, 2008).
Anderson, K. C. New insights into therapeutic targets in myeloma. ASH Education Book 2011, 184–190 (2011).
Walker, B. A. et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood 117, 553–562 (2011). The first report of global methylation changes from MGUS to PCL and within the myeloma cytogenetic subgroups.
Rasmussen, T. et al. Identification of translocation products but not K-RAS mutations in memory B cells from multiple myeloma patients. Haematologica 95, 1730–1737 (2010).
Altieri, A., Chen, B., Bermejo, J. L., Castro, F. & Hemminki, K. Familial risks and temporal incidence trends of multiple myeloma. Eur. J. Cancer 42, 1661–1670 (2006).
Broderick, P. et al. Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nature Genet. 44, 58–61 (2012). The first GWAS to identify myeloma disease susceptibility loci.
Bergsagel, P. L. et al. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 106, 296–303 (2005).
Chesi, M. et al. Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood 88, 674–681 (1996).
Shaughnessy, J. Jr et al. Cyclin D3 at 6p21 is dysregulated by recurrent chromosomal translocations to immunoglobulin loci in multiple myeloma. Blood 98, 217–223 (2001).
Hurt, E. M. et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 5, 191–199 (2004).
Shou, Y. et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc. Natl Acad. Sci. USA 97, 228–233 (2000).
Avet-Loiseau, H. et al. Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood 98, 3082–3086 (2001).
Chesi, M. et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 13, 167–180 (2008).
Annunziata, C. M. et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007).
Carrasco, R. et al. Comprehensive genome-wide profile of regional gains and losses in multiple myeloma using array-CGH: The 1q21 amplification and potential role of the BCL-9 gene in multiple myeloma pathogenesis. Blood (ASH Annual Meeting Abstracts) 104, 785 (2004).
Keats, J. J. et al. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell 12, 131–144 (2007). References 47 and 49 highlight the importance of NF-κB signalling in myeloma and the role that deregulation of the pathway has in myeloma pathogenesis.
Dickens, N. J. et al. Homozygous deletion mapping in myeloma samples identifies genes and an expression signature relevant to pathogenesis and outcome. Clin. Cancer Res. 16, 1856–1864 (2010).
Boyd, K. D. et al. Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clin. Cancer Res. 17, 7776–7784 (2011).
Jenner, M. W. et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood 110, 3291–3300 (2007).
Leone, P. E. et al. Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin. Cancer Res. 14, 6033–6041 (2008).
Gonzalez-Paz, N. et al. Tumor suppressor p16 methylation in multiple myeloma: biological and clinical implications. Blood 109, 1228–1232 (2007).
Dib, A., Barlogie, B., Shaughnessy, J. D. Jr & Kuehl, W. M. Methylation and expression of the p16INK4A tumor suppressor gene in multiple myeloma. Blood 109, 1337–1338 (2007).
Davis, R. E., Brown, K. D., Siebenlist, U. & Staudt, L. M. Constitutive nuclear factor-κB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 194, 1861–1874 (2001).
Onodera, N., McCabe, N. R. & Rubin, C. M. Formation of a hyperdiploid karyotype in childhood acute lymphoblastic leukemia. Blood 80, 203–208 (1992).
Shaughnessy, J. Amplification and overexpression of CKS1B at chromosome band 1q21 is associated with reduced levels of p27Kip1 and an aggressive clinical course in multiple myeloma. Hematology. 10 (Suppl. 1), 117–126 (2005).
Ley, T. J. et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456, 66–72 (2008).
Lee, W. et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 465, 473–477 (2010).
Tiacci, E. et al. BRAF mutations in hairy-cell leukemia. N. Engl. J. Med. 364, 2305–2315 (2011).
Aronson, L. I. et al. Autophagy is a key myeloma survival pathway that can be manipulated therapeutically to enhance apoptosis. ASH Annual Meeting Abstracts 116, 4083 (2010).
Boyd, K. D. et al. High expression levels of the mammalian target of rapamycin inhibitor DEPTOR are predictive of response to thalidomide in myeloma. Leuk. Lymphoma 51, 2126–2129 (2010).
Fazzari, M. J. & Greally, J. M. Epigenomics: beyond CpG islands. Nature Rev. Genet. 5, 446–455 (2004).
Chi, P., Allis, C. D. & Wang, G. G. Covalent histone modifications — miswritten, misinterpreted and mis-erased in human cancers. Nature Rev. Cancer 10, 457–469 (2010).
Cedar, H. & Bergman, Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Rev. Genet. 10, 295–304 (2009).
Nimura, K. et al. A histone H3 lysine 36 trimethyltransferase links Nkx2–5 to Wolf-Hirschhorn syndrome. Nature 460, 287–291 (2009).
Kim, J. Y. et al. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol. Cell Biol. 28, 2023–2034 (2008).
Marango, J. et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood 111, 3145–3154 (2008).
Kuo, A. J. et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 44, 609–620 (2011).
Martinez-Garcia, E. et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 117, 211–220 (2011).
Brito, J. L. et al. MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells. Haematologica 94, 78–86 (2009).
Pei, H. et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470, 124–128 (2011).
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genet. 41, 521–523 (2009).
Avet-Loiseau, H. et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p). J. Clin. Oncol. 28, 4630–4634 (2010).
Lode, L. et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica 95, 1973–1976 (2010).
Boyd, K. D. et al. The clinical impact and molecular biology of del(17p) in multiple myeloma treated with conventional or thalidomide-based therapy. Genes Chromosomes Cancer 50, 765–774 (2011).
Pichiorri, F. et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl Acad. Sci. USA 105, 12885–12890 (2008).
Pichiorri, F. et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 18, 367–381 (2010).
Zenz, T. et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood 114, 2589–2597 (2009).
Benetatos, L. & Vartholomatos, G. Deregulated microRNAs in multiple myeloma. Cancer 118, 878–887 (2012).
Chang, T. C. et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genet. 40, 43–50 (2008).
Boyd, K. D. et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia 26, 349–355 (2012). Evidence is presented highlighting the importance of combining cytogenetic abnormalities with other disease markers to accurately predict therapeutic response.
Ross, F. M. et al. The t(14;20) is a poor prognostic factor in myeloma but is associated with long-term stable disease in monoclonal gammopathies of undetermined significance. Haematologica 95, 1221–1225 (2010).
Chauhan, D. et al. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-κB. Blood 87, 1104–1112 (1996).
Moreaux, J. et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood 103, 3148–3157 (2004).
Neri, P. et al. Integrin β7-mediated regulation of multiple myeloma cell adhesion, migration, and invasion. Blood 117, 6202–6213 (2011).
Annunziata, C. M. et al. A mechanistic rationale for MEK inhibitor therapy in myeloma based on blockade of MAF oncogene expression. Blood 117, 2396–2404 (2011).
Wu, P. et al. A gene expression-based predictor for myeloma patients at high risk of developing bone disease on bisphosphonate treatment. Clin. Cancer Res. 17, 6347–6355 (2011).
Bezieau, S. et al. High incidence of N and K-Ras activating mutations in multiple myeloma and primary plasma cell leukemia at diagnosis. Hum. Mutat. 18, 212–224 (2001).
Braga, V. M., Betson, M., Li, X. & Lamarche-Vane, N. Activation of the small GTPase Rac is sufficient to disrupt cadherin-dependent cell-cell adhesion in normal human keratinocytes. Mol. Biol. Cell 11, 3703–3721 (2000).
Allan, V. J. Cytoplasmic dynein. Biochem. Soc. Trans. 39, 1169–1178 (2011).
Morgan, G. J. et al. First-line treatment with zoledronic acid as compared with clodronic acid in multiple myeloma (MRC Myeloma IX): a randomised controlled trial. Lancet 376, 1989–1999 (2010).
Tian, E. et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N. Engl. J. Med. 349, 2483–2494 (2003).
Vallet, S. et al. Activin A promotes multiple myeloma-induced osteolysis and is a promising target for myeloma bone disease. Proc. Natl Acad. Sci. USA 107, 5124–5129 (2010).
Avet-Loiseau, H. et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood 109, 3489–3495 (2007).
Avet-Loiseau, H. et al. Prognostic significance of copy-number alterations in multiple myeloma. J. Clin. Oncol. 27, 4585–4590 (2009).
Morgan, G. J. et al. Cyclophosphamide, thalidomide, and dexamethasone (CTD) as initial therapy for patients with multiple myeloma unsuitable for autologous transplantation. Blood 118, 1231–1238 (2011).
Paiva, B. et al. High-risk cytogenetics and persistent minimal residual disease by multiparameter flow cytometry predict unsustained complete response after autologous stem cell transplantation in multiple myeloma. Blood 119, 687–691 (2012).
Munshi, N. C. et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood 117, 4696–4700 (2011). An international consensus statement discussing the clinical impact of cytogenetic abnormalities.
Broyl, A. et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood 116, 2543–2553 (2010).
Zhan, F. et al. Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood 101, 1128–1140 (2003).
Nair, B. et al. Gene expression profiling of plasma cells at myeloma relapse from tandem transplantation trial Total Therapy 2 predicts subsequent survival. Blood 113, 6572–6575 (2009).
Chng, W. J., Glebov, O., Bergsagel, P. L. & Kuehl, W. M. Genetic events in the pathogenesis of multiple myeloma. Best Pract. Res. Clin. Haematol. 20, 571–596 (2007).
Kassambara, A. et al. Genes with a spike expression are clustered in chromosome (sub)bands and spike (sub)bands have a powerful prognostic value in patients with multiple myeloma. Haematologica 18 Nov 2011 (doi:10.3324/haematol.2011.046821).
Zhan, F. et al. The molecular classification of multiple myeloma. Blood 108, 2020–2028 (2006).
Shaughnessy, J. D. Jr et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 109, 2276–2284 (2007).
Morgan, G. J. et al. The role of maintenance thalidomide therapy in multiple myeloma: MRC Myeloma IX results and meta-analysis. Blood 119, 7–15 (2012).
Neben, K. et al. Administration of bortezomib before and after autologous stem-cell transplantation improves outcome in multiple myeloma patients with deletion 17p. Blood 119, 940–948 (2012).
Stewart, A. K. Whole genome sequencing in high-risk myeloma. 13th Annual International Myeloma Workshop 2011 [online] (2011).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Rettig, M. B. et al. VH gene usage in multiple myeloma: complete absence of the VH4.21 (VH4-34) gene. Blood 87, 2846–2852 (1996).
Nobuyoshi, M. et al. Increased expression of the c-myc gene may be related to the aggressive transformation of human myeloma cells. Br. J. Haematol. 77, 523–528 (1991).
Perez-Rosado, A. et al. BCL6 represses NFκB activity in diffuse large B-cell lymphomas. J. Pathol. 214, 498–507 (2008).
Acknowledgements
We would like to acknowledge Myeloma UK for programme funding. F.E.D. is a Cancer Research UK Senior Cancer Fellow. In addition, we would also like to thank M. Greaves, L. Melchor, M. Kaiser and D. Gonzalez for their helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
DATABASES
FURTHER INFORMATION
Glossary
- Paraproteinaemias
-
The clinical diseases that produce a paraprotein (immunoglobulin from a single clone that is present at high levels in the serum) and include myeloma, monoclonal gammopathy of undetermined significance (MGUS) and Waldenstrom's macroglobulinaemia (WM).
- Plasma cell
-
A terminally differentiated quiescent B cell that secretes large amounts of antibodies.
- Germinal centre
-
The site within a lymph node where B cells undergo affinity maturation of their antibody through somatic hypermutation (SHM). During this process the B cells rapidly proliferate and differentiate and are selected for increasingly specific antibodies against the antigen.
- Class switch recombination
-
(CSR). A region-specific deletional recombination reaction that replaces one switch region with another. This allows the production of different immunoglobulin isotypes.
- Immunoglobulin isotypes
-
Immunoglobulin (Ig) isotypes are defined by the heavy and light chain locus constant regions used by the Ig. These constant regions are encoded within the IGH@ locus (heavy chain) on chromosome 14 or the IGK@ and IGL@ loci (light chain) on chromosomes 2 and 22, respectively. There are nine heavy chain isotypes: IgM, IgD, IgE, IgA1, IgA2, IgG1, IgG2, IgG3 and IgG4. Two light chain isotypes exist, kappa and lambda.
- Centroblast
-
A lymphocyte that has a large, non-cleaved nucleus. B cells that are proliferating and undergoing affinity maturation in response to stimulus, within the germinal centre, are termed centroblasts. These cells mature to centrocytes within the germinal centre before they leave to become plasmablasts.
- Unfolded protein response
-
(UPR). This is a cellular stress pathway that is activated in response to an accumulation of unfolded or misfolded proteins. In myeloma plasma cells this pathway is crucial for dealing with the large quantities of immunoglobulin (or paraprotein) that are produced by the cells. Disruption of this pathway in plasma cells can lead to apoptosis.
- Serological memory
-
The presence of immunoglobulin in the serum, usually from mature B cells that have undergone somatic hypermutation and class switch recombination in a germinal centre.
Rights and permissions
About this article
Cite this article
Morgan, G., Walker, B. & Davies, F. The genetic architecture of multiple myeloma. Nat Rev Cancer 12, 335–348 (2012). https://doi.org/10.1038/nrc3257
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3257
This article is cited by
-
Expression of integrin β-7 is epigenetically enhanced in multiple myeloma subgroups with high-risk cytogenetics
Clinical Epigenetics (2023)
-
A CRISPR interference strategy for gene expression silencing in multiple myeloma cell lines
Journal of Biological Engineering (2023)
-
FAM72D in plasma cell myeloma: a friend or enemy
Egyptian Journal of Medical Human Genetics (2023)
-
Integrative analysis of the prognostic value and immune microenvironment of mitophagy-related signature for multiple myeloma
BMC Cancer (2023)
-
Circulating tumour DNA analysis predicts relapse and improves risk stratification in primary refractory multiple myeloma
Blood Cancer Journal (2023)