Key Points
-
Integrated screens of DNA copy number and gene expression in human cancers using microarray platforms have accelerated the rate of discovery of amplified and overexpressed genes. However, the biological importance of most of the genes identified in such studies remains unclear.
-
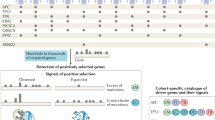
Amplification events often include multiple genes, so consideration of the pattern of genetic alteration alone is usually insufficient to identify which gene in an amplicon is being selected for owing to its contribution to oncogenesis. Supplementary datasets are usually required, including physical mapping, determination of overexpression, correlation with clinical outcome, biological investigations of function and, in some cases, efficacy of drugs targeted against the encoded overexpressed proteins.
-
In this Analysis we propose a weight-of-evidence based classification system for identifying individual genes in amplified regions that are selected for in an amplicon and so contribute to cancer development. The proposed classification scheme takes into account the complex and sometimes distinct datasets that are available for different amplicons in cancer.
-
Using this classification scheme in a census of the published literature, we have identified 77 genes for which there is evidence of involvement in human cancer development.
-
The 77 genes were divided into three classes based on the weight of supporting evidence. We consider that for class II (12 genes), and class I (3 genes) the evidence is sufficiently strong for their inclusion in the census of human cancer genes.
-
Linking newly generated integrated datasets to supporting evidence using the criteria outlined in this Analysis will aid in the future identification of amplified and overexpressed genes that contribute to cancer development.
Abstract
Integrated genome-wide screens of DNA copy number and gene expression in human cancers have accelerated the rate of discovery of amplified and overexpressed genes. However, the biological importance of most of the genes identified in such studies remains unclear. In this Analysis, we propose a weight-of-evidence based classification system for identifying individual genes in amplified regions that are selected for during tumour development. In a census of the published literature we have identified 77 genes for which there is good evidence of involvement in the development of human cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Futreal PA. et al. A census of human cancer genes. Nature Rev. Cancer 4, 177–183 (2004).
Bignell GR. et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 17, 1296–1303 (2007).
Savelyeva L. & Schwab M. Amplification of oncogenes revisited: from expression profiling to clinical application. Cancer Lett. 167, 115–123 (2001).
Myllykangas S. & Knuutila S. Manifestation, mechanisms and mysteries of gene amplifications. Cancer Lett. 232, 79–89 (2006).
Schwab M. Oncogene amplification in solid tumors. Semin. Cancer Biol. 9, 319–325 (1999).
Natrajan R. et al. Amplification and overexpression of CACNA1E correlates with relapse in favorable histology Wilms' tumors. Clin. Cancer Res. 12, 7284–7293 (2006).
McIntyre A. et al. Amplification and overexpression of the KIT gene is associated with progression in the seminoma subtype of testicular germ cell tumors of adolescents and adults. Cancer Res. 65, 8085–8089 (2005).
Fix A. et al. Characterization of amplicons in neuroblastoma: high-resolution mapping using DNA microarrays, relationship with outcome, and identification of overexpressed genes. Genes Chromosom. Cancer 47, 819–834 (2008).
Weber A, Imisch P, Bergmann E & Christiansen H. Coamplification of DDX1 correlates with an improved survival probability in children with MYCN-amplified human neuroblastoma. J. Clin. Oncol. 22, 2681–2690 (2004).
Kaneko S., Ohira M, Nakamura Y, Isogai E, Nakagawara A & Kaneko M. Relationship of DDX1 and NAG gene amplification/overexpression to the prognosis of patients with MYCN-amplified neuroblastoma. J. Cancer Res. Clin. Oncol. 133, 185–192 (2007).
De Preter K. et al. No evidence for correlation of DDX1 gene amplification with improved survival probability in patients with MYCN-amplified neuroblastomas. J. Clin. Oncol. 23, 3167–3168 (2005).
Rodriguez S. et al. Expression profile of genes from 12p in testicular germ cell tumors of adolescents and adults associated with i(12p) and amplification at 12p11.2-p12.1. Oncogene 22, 1880–221891 (2003).
Zafarana G. et al. 12p-amplicon structure analysis in testicular germ cell tumors of adolescents and adults by array CGH. Oncogene 22, 7695–7701 (2003).
Bourdon V. et al. Genomic and expression analysis of the 12p11-p12 amplicon using EST arrays identifies two novel amplified and overexpressed genes. Cancer Res. 62, 6218–6223 (2002).
Ormandy CJ, Musgrove EA, Hui R., Daly RJ & Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res. Treat. 78, 323–335 (2003).
Albertson DG. Gene amplification in cancer. Trends Genet. 22, 447–455 (2006).
Cordon-Cardo C. et al. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 54, 794–799 (1994).
Hayward NK. Genetics of melanoma predisposition. Oncogene 22, 3053–3062 (2003).
Feber A. et al. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene 23, 1627–1630 (2004).
Oeggerli M. et al. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene 23, 5616–5623 (2004).
Kao J. & Pollack JR. RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes. Genes Chromosom. Cancer 45, 761–769 (2006).
Comoglio PM, Giordano S. & Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nature Rev. Drug Discov. 7, 504–516 (2008).
Zender L. et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell 125, 1253–1267 (2006).
Vogel CL. et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol. 20, 719–726 (2002).
Mass RD. et al. Evaluation of clinical outcomes according to HER2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with trastuzumab. Clin. Breast Cancer 6, 240–246 (2005).
Ray ME. et al. Genomic and expression analysis of the 8p11–12 amplicon in human breast cancer cell lines. Cancer Res. 64, 40–47 (2004).
Garcia MJ. et al. A 1 Mb minimal amplicon at 8p11–12 in breast cancer identifies new candidate oncogenes. Oncogene 24, 5235–5245 (2005).
Reis-Filho JS. et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin. Cancer Res. 12, 6652–6662 (2006).
Campbell PJ. et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. 40, 722–729 (2008).
Sugawa N., Ekstrand AJ, James CD & Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc. Natl Acad. Sci. USA 87, 8602–8606 (1990).
Weber-Hall S. et al. Novel formation and amplification of the PAX7-FKHR fusion gene in a case of alveolar rhabdomyosarcoma. Genes Chromosom. Cancer 17, 7–13 (1996).
Nikolsky Y. et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res. 68, 9532–9540 (2008).
Acknowledgements
The authors thank the National Cancer Research Institute, the Grand Charity of Freemasons, the Prostate Cancer Charity, the Bob Champion Cancer Trust, the Prostate Cancer Research Foundation, and the Wellcome Trust for Support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (text)
A survey of amplified and overexpressed genes in human cancer (PDF 2900 kb)
Supplementary information S2 (table)
Amplified and overexpressed genes in human cancer (XLS 48 kb)
Supplementary information S3 (text)
A Review of Integrated datasets (PDF 342 kb)
Related links
Related links
FURTHER INFORMATION
Cancer Genetics Programme at the sanger Centre
Institute for Cancer Research Amplified and Overexpressed Genes in Cancer (AOGIC) dataset
Rights and permissions
About this article
Cite this article
Santarius, T., Shipley, J., Brewer, D. et al. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer 10, 59–64 (2010). https://doi.org/10.1038/nrc2771
Issue Date:
DOI: https://doi.org/10.1038/nrc2771
This article is cited by
-
Clinical value of Cyclin D1 and P21 in the differential diagnosis of papillary thyroid carcinoma
Diagnostic Pathology (2023)
-
NCBP2 and TFRC are novel prognostic biomarkers in oral squamous cell carcinoma
Cancer Gene Therapy (2023)
-
Activation of invasion by oncogenic reprogramming of cholesterol metabolism via increased NPC1 expression and macropinocytosis
Oncogene (2023)
-
Pan-cancer analysis of genomic and transcriptomic data reveals the prognostic relevance of human proteasome genes in different cancer types
BMC Cancer (2022)
-
CPT1A-mediated fatty acid oxidation promotes cell proliferation via nucleoside metabolism in nasopharyngeal carcinoma
Cell Death & Disease (2022)