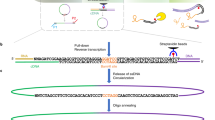
Abstract
The identification of sites where RNA-binding proteins (RNABPs) interact with target RNAs opens the door to understanding the vast complexity of RNA regulation. UV cross-linking and immunoprecipitation (CLIP) is a transformative technology in which RNAs purified from in vivo cross-linked RNA-protein complexes are sequenced to reveal footprints of RNABP:RNA contacts. CLIP combined with high-throughput sequencing (HITS-CLIP) is a generalizable strategy to produce transcriptome-wide maps of RNA binding with higher accuracy and resolution than standard RNA immunoprecipitation (RIP) profiling or purely computational approaches. The application of CLIP to Argonaute proteins has expanded the utility of this approach to mapping binding sites for microRNAs and other small regulatory RNAs. Finally, recent advances in data analysis take advantage of cross-link–induced mutation sites (CIMS) to refine RNA-binding maps to single-nucleotide resolution. Once IP conditions are established, HITS-CLIP takes ∼8 d to prepare RNA for sequencing. Established pipelines for data analysis, including those for CIMS, take 3–4 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
24 February 2016
In the version of this article initially published, units were incorrectly reported in Step 2 of the Procedure. They have been changed from 'mJ cm2' to 'mJ per cm2' in four instances. In addition, units that previously read '1 = 0.1 J per m2' have been changed to '1 = 1 J per m2'. These errors have been corrected in the HTML and PDF versions of the article.
References
Czech, B. & Hannon, G.J. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 12, 19–31 (2011).
Joshua-Tor, L. & Hannon, G.J. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb. Perspect. Biol. 3, a003772 (2011).
Fabian, M.R., Sonenberg, N. & Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 (2010).
Chi, S.W., Zang, J.B., Mele, A. & Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486 (2009).
Gaken, J. et al. A functional assay for microRNA target identification and validation. Nucleic Acids Res. 40, e75 (2012).
Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Chi, S.W., Hannon, G.J. & Darnell, R.B. An alternative mode of microRNA target recognition. Nat. Struct. Mol. Biol. 19, 321–327 (2012).
Ule, J., Jensen, K., Mele, A. & Darnell, R.B. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37, 376–386 (2005).
Zhang, C. & Darnell, R.B. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotechnol. 29, 607–614 (2011).
Granneman, S., Kudla, G., Petfalski, E. & Tollervey, D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. USA 106, 9613–9618 (2009).
Ule, J. et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 37, 844–852 (2005).
Ule, J. & Darnell, R.B. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 16, 102–110 (2006).
Ule, J. et al. An RNA map predicting Nova-dependent splicing regulation. Nature 444, 580–586 (2006).
Darnell, R.B. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev. RNA 1, 266–286 (2010).
Urlaub, H., Hartmuth, K. & Luhrmann, R. A two-tracked approach to analyze RNA-protein cross-linking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods 26, 170–181 (2002).
Ule, J. et al. CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 (2003).
Licatalosi, D.D. et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469 (2008).
Licatalosi, D.D. & Darnell, R.B. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 11, 75–87 (2010).
Sanford, J.R. et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 19, 381–394 (2009).
Tollervey, J.R. et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 (2011).
Darnell, J.C. et al. FMRP Stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011).
Racca, C. et al. The neuronal splicing factor Nova co-localizes with target RNAs in the dendrite. Front Neural Circuits 4, 5 (2010).
Yano, M., Hayakawa-Yano, Y., Mele, A. & Darnell, R.B. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron 66, 848–858 (2010).
Xue, Y. et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell 36, 996–1006 (2009).
Licatalosi, D.D. et al. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 26, 1626–1642 (2012).
Mukherjee, N. et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 43, 327–339 (2011).
Lebedeva, S. et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43, 340–352 (2011).
Ince-Dunn, G. et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron 75, 1067–1080 (2012).
Wang, Z. et al. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 8, 1–16 (2010).
Polymenidou, M. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 (2011).
Konig, J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909–915 (2010).
Yeo, G.W. et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16, 130–137 (2009).
Charizanis, K. et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 75, 437–450 (2012).
Leung, A.K. et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 18, 237–244 (2011).
Loeb, G.B. et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 48, 760–770 (2012).
Riley, K.J. et al. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 31, 2207–2221 (2012).
Zisoulis, D.G. et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 17, 173–179 (2010).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Mili, S. & Steitz, J.A. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10, 1692–1694 (2004).
Kishore, S. et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 8, 559–564 (2011).
Kemeny-Beke, A. et al. Antiproliferative effect of 4-thiouridylate on OCM-1 uveal melanoma cells. Eur. J. Ophthalmol. 16, 680–685 (2006).
Burger, K. et al. 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 10 (2013).
Khan, A.A. et al. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 27, 549–555 (2009).
Hafner, M. et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 17, 1697–1712 (2011).
Zhuang, F., Fuchs, R.T., Sun, Z., Zheng, Y. & Robb, G.B. Structural bias in T4 RNA ligase-mediated 3-adapter ligation. Nucleic Acids Res. 40, e54 (2012).
Harlow, E. & Lane, D. Antibodies, A Laboratory Manual (Cold Spring Harbor Laboratory Press, 1988).
Darnell, J.C., Mele, A. & Darnell, R.B. Mapping of in vivo RNA binding sites by UV-crosslinking immunoprecipitation (CLIP). In Molecular Cloning: A Laboratory Manual (eds Green, M. & Sambrook, J.) (Cold Spring Harbor Laboratory Press, 2012).
Parker, J.S. How to slice: snapshots of Argonaute in action. Silence 1, 3 (2010).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Granneman, S., Petfalski, E., Swiatkowska, A. & Tollervey, D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 29, 2026–2036 (2010).
Bohnsack, M.T. et al. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell 36, 583–592 (2009).
Wang, E.T. et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150, 710–724 (2012).
Cho, J. et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151, 765–777 (2012).
Rhead, B. et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 38, D613–9 (2010).
Bailey, T.L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994).
Tempel, S. Using and understanding RepeatMasker. Methods Mol. Biol. 859, 29–51 (2012).
Sugimoto, Y. et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 13, R67 (2012).
Cooper, G.M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 15, 901–913 (2005).
Acknowledgements
We are indebted to Z. Mourelatos for sharing 2A8 antibody. We thank S. Dewell, C. Zhao and the entire staff of the Rockefeller University Genomics Resource Center for their expertise and support. We are grateful to sources of support for M.J.M. (Jane Coffin Childs Fund for Medical Research), C.Z. (US National Institutes of Health (NIH) grant no. K99/R00GM95713 and Simons Foundation grant no. 297990) and R.B.D. (grant nos. NIH NS081706 and NIH NS34389, the Simons Foundation and the Howard Hughes Medical Institute).
Author information
Authors and Affiliations
Contributions
M.J.M. wrote the sections of the manuscript pertaining to the experimental protocol and produced the data in Figure 2. C.Z. developed the methods and wrote manuscript sections pertaining to the bioinformatic protocol, including the analyses in Figures 4,5,6,7. E.C.G. produced the data in Figure 3a. A.M. produced the data in Figure 3b. J.C.D. contributed to writing sections of the experimental protocol. R.B.D. contributed to writing all sections and directed the development of all experimental and bioinformatic methods described here.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Moore, M., Zhang, C., Gantman, E. et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc 9, 263–293 (2014). https://doi.org/10.1038/nprot.2014.012
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.012
This article is cited by
-
An mRNA processing pathway suppresses metastasis by governing translational control from the nucleus
Nature Cell Biology (2023)
-
LncRNA BCAN-AS1 stabilizes c-Myc via N6-methyladenosine-mediated binding with SNIP1 to promote pancreatic cancer
Cell Death & Differentiation (2023)
-
CSTF2 mediated mRNA N6-methyladenosine modification drives pancreatic ductal adenocarcinoma m6A subtypes
Nature Communications (2023)
-
A first-in-class fully modified version of miR-34a with outstanding stability, activity, and anti-tumor efficacy
Oncogene (2023)
-
The m6A reading protein YTHDF3 potentiates tumorigenicity of cancer stem-like cells in ocular melanoma through facilitating CTNNB1 translation
Oncogene (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.