Abstract
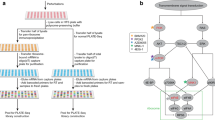
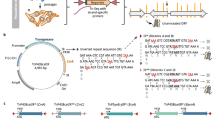
Parallel analysis of translated open reading frames (ORFs) (PLATO) can be used for the unbiased discovery of interactions between full-length proteins encoded by a library of 'prey' ORFs and surface-immobilized 'bait' antibodies, polypeptides or small-molecular-weight compounds. PLATO uses ribosome display (RD) to link ORF-derived mRNA molecules to the proteins they encode, and recovered mRNA from affinity enrichment is subjected to analysis using massively parallel DNA sequencing. Compared with alternative in vitro methods, PLATO provides several advantages including library size and cost. A unique advantage of PLATO is that an alternative reverse transcription–quantitative PCR (RT-qPCR) protocol can be used to test binding of specific, individual proteins. To illustrate a typical experimental workflow, we demonstrate PLATO for the identification of the immune target of serum antibodies from patients with inclusion body myositis (IBM). Beginning with an ORFeome library in an RD vector, the protocol can produce samples for deep sequencing or RT-qPCR within 4 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Zhu, J. et al. Protein interaction discovery using parallel analysis of translated ORFs (PLATO). Nat. Biotechnol. 31, 331–334 (2013).
Hanes, J. & Pluckthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 94, 4937–4942 (1997).
Takahashi, T.T., Austin, R.J. & Roberts, R.W. mRNA display: ligand discovery, interaction analysis and beyond. Trends Biochem. Sci. 28, 159–165 (2003).
Walhout, A.J. et al. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328, 575–592 (2000).
Brasch, M.A., Hartley, J.L. & Vidal, M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 14, 2001–2009 (2004).
Temple, G. et al. The completion of the mammalian gene collection (MGC). Genome Res. 19, 2324–2333 (2009).
Yang, X. et al. A public genome-scale lentiviral expression library of human ORFs. Nat. Methods 8, 659–661 (2011).
Brandner, C.J. et al. The ORFeome of Staphylococcus aureus v 1.1. BMC Genomics 9, 321 (2008).
D'Angelo, S. et al. Filtering 'genic' open reading frames from genomic DNA samples for advanced annotation. BMC Genomics 12 (suppl. 1), S5 (2011).
Hartley, J.L., Temple, G.F. & Brasch, M.A. DNA cloning using in vitro site-specific recombination. Genome Res 10, 1788–1795 (2000).
Katzen, F. Gateway recombinational cloning: a biological operating system. Exp. Opin. Drug Discov. 2, 571–589 (2007).
Lamesch, P. et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics 89, 307–315 (2007).
Yu, H. et al. Next-generation sequencing to generate interactome datasets. Nat. Methods 8, 478–480 (2011).
Dreier, B. & Pluckthun, A. Rapid selection of high-affinity binders using ribosome display. Methods Mol. Biol. 805, 261–286 (2012).
Hajnsdorf, E., Braun, F., Haugel-Nielsen, J., Le Derout, J. & Regnier, P. Multiple degradation pathways of the rpsO mRNA of Escherichia coli. RNase E interacts with the 5′ and 3′ extremities of the primary transcript. Biochimie 78, 416–424 (1996).
Carlson, E.D., Gan, R., Hodgman, C.E. & Jewett, M.C. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 30, 1185–1194 (2012).
Goshima, N. et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 5, 1011–1017 (2008).
He, M. & Taussig, M.J. Eukaryotic ribosome display with in situ DNA recovery. Nat. Methods 4, 281–288 (2007).
Dabney, J. & Meyer, M. Length and GC-biases during sequencing library amplification: a comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. BioTechniques 52, 87–94 (2012).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Fonseca, N.A., Rung, J., Brazma, A. & Marioni, J.C. Tools for mapping high-throughput sequencing data. Bioinformatics 28, 3169–3177 (2012).
Somwar, R. et al. Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 108, 16375–16380 (2011).
Larman, H.B. et al. Cytosolic 5′-nucleotidase 1A autoimmunity in sporadic inclusion body myositis. Ann. Neurol. 73, 408–418 (2013).
Pluk, H. et al. Autoantibodies to cytosolic 5′-nucleotidase 1A in inclusion body myositis. Ann. Neurol. 73, 397–407 (2013).
Acknowledgements
IBM patient sera were kindly provided by Stephen A. Greenberg (Brigham and Women's Hospital). The ORFeome library v5.1 was provided by the Center for Cancer Systems Biology (CCSB) at the Dana-Farber Cancer Institute (Boston, Massachusetts). This work was supported in part by a grant from the Leona M. and Harry B. Helmsley Charitable Trust. S.J.E. is an investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
H.B.L., S.J.E. and J.Z. developed the protocol. H.B.L. and J.Z. wrote the paper. J.Z. performed the PLATO screen with IBM samples. A.C.L. performed the immunofluorescence.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Larman, H., Liang, A., Elledge, S. et al. Discovery of protein interactions using parallel analysis of translated ORFs (PLATO). Nat Protoc 9, 90–103 (2014). https://doi.org/10.1038/nprot.2013.167
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.167
This article is cited by
-
High throughput protease profiling comprehensively defines active site specificity for thrombin and ADAMTS13
Scientific Reports (2018)
-
PhIP-Seq characterization of serum antibodies using oligonucleotide-encoded peptidomes
Nature Protocols (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.