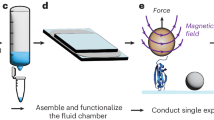
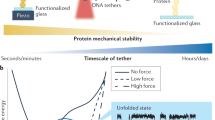
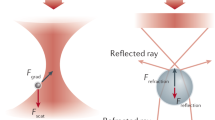
Abstract
Here we describe a protocol for using force-clamp spectroscopy to precisely quantify the effect of force on biochemical reactions. A calibrated force is used to control the exposure of reactive sites in a single polyprotein substrate composed of repeated domains. The use of polyproteins allows the identification of successful single-molecule recordings from unambiguous mechanical unfolding fingerprints. Biochemical reactions are then measured directly by detecting the length changes of the substrate held at a constant force. We present the layout of a force-clamp spectrometer along with protocols to design and conduct experiments. These experiments measure reaction kinetics as a function of applied force. We show sample data of the force dependency of two different reactions, protein unfolding and disulfide reduction. These data, which can be acquired in just a few days, reveal mechanistic details of the reactions that currently cannot be resolved by any other technique.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Rief, M., Gautel, M., Oesterhelt, F., Fernandez, J.M. & Gaub, H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 (1997).
Oberhauser, A.F., Marszalek, P.E., Erickson, H.P. & Fernandez, J.M. The molecular elasticity of the extracellular matrix protein tenascin. Nature 393, 181–185 (1998).
Rief, M., Oesterhelt, F., Heymann, B. & Gaub, H.E. Single-molecule force spectroscopy on polysaccharides by atomic force microscopy. Science 275, 1295–1297 (1997).
Smith, S.B., Cui, Y.J. & Bustamante, C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799 (1996).
Carrion-Vazquez, M. et al. Mechanical design of proteins studied by single-molecule force spectroscopy and protein engineering. Prog. Biophys. Mol. Biol. 74, 63–91 (2000).
Oberhauser, A.F., Badilla-Fernandez, C., Carrion-Vazquez, M. & Fernandez, J.M. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 319, 433–447 (2002).
Li, H. et al. Reverse engineering of the giant muscle protein titin. Nature 418, 998–1002 (2002).
Carrion-Vazquez, M. et al. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 10, 738–743 (2003).
Brockwell, D.J. et al. Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat. Struct. Biol. 10, 731–737 (2003).
Dietz, H. & Rief, M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA 101, 16192–16197 (2004).
Iozzi, M.F., Helgaker, T. & Uggerud, E. Influence of external force on properties and reactivity of disulfide bonds. J. Phys. Chem. A 115, 2308–2315 (2011).
Ainavarapu, S.R. et al. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys. J. 92, 225–233 (2007).
Wiita, A.P., Ainavarapu, S.R., Huang, H.H. & Fernandez, J.M. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc. Natl. Acad. Sci. USA 103, 7222–7227 (2006).
Oberhauser, A.F., Hansma, P.K., Carrion-Vazquez, M. & Fernandez, J.M. Stepwise unfolding of titin under force-clamp atomic force microscopy. Proc. Natl. Acad. Sci. USA 98, 468–472 (2001).
Schlierf, M., Li, H. & Fernandez, J.M. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl. Acad. Sci. USA 101, 7299–7304 (2004).
Wiita, A.P. et al. Probing the chemistry of thioredoxin catalysis with force. Nature 450, 124–127 (2007).
Kuo, T.L. et al. Probing static disorder in Arrhenius kinetics by single-molecule force spectroscopy. Proc. Natl. Acad. Sci. USA 107, 11336–11340 (2010).
Garcia-Manyes, S., Dougan, L. & Fernandez, J.M. Osmolyte-induced separation of the mechanical folding phases of ubiquitin. Proc. Natl. Acad. Sci. USA 106, 10540–10545 (2009).
Popa, I., Fernandez, J.M. & Garcia-Manyes, S. Direct quantification of the attempt frequency determining the mechanical unfolding of ubiquitin protein. J. Biol. Chem. 286, 31072–31079 (2011).
Garcia-Manyes, S., Dougan, L., Badilla, C.L., Brujic, J. & Fernandez, J.M. Direct observation of an ensemble of stable collapsed states in the mechanical folding of ubiquitin. Proc. Natl. Acad. Sci. USA 106, 10534–10539 (2009).
Berkovich, R. et al. Rate limit of protein elastic response is tether dependent. Proc. Natl. Acad. Sci. USA 109, 14416–14421 (2012).
Garcia-Manyes, S., Brujic, J., Badilla, C.L. & Fernandez, J.M. Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin. Biophys. J. 93, 2436–2446 (2007).
Walther, K.A. et al. Signatures of hydrophobic collapse in extended proteins captured with force spectroscopy. Proc. Natl. Acad. Sci. USA 104, 7916–7921 (2007).
Garcia-Manyes, S., Liang, J., Szoszkiewicz, R., Kuo, T.L. & Fernandez, J.M. Force-activated reactivity switch in a bimolecular chemical reaction. Nat. Chem. 1, 236–242 (2009).
Ainavarapu, S.R.K., Wiita, A.P., Dougan, L., Uggerud, E. & Fernandez, J.M. Single-molecule force spectroscopy measurements of bond elongation during a bimolecular reaction. J. Am. Chem. Soc. 130, 6479–6487 (2008).
Liang, J. & Fernandez, J.M. Kinetic measurements on single-molecule disulfide bond cleavage. J. Am. Chem. Soc. 133, 3528–3534 (2011).
Perez-Jimenez, R. et al. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat. Struct. Mol. Biol. 18, U592–U599 (2011).
Perez-Jimenez, R. et al. Diversity of chemical mechanisms in thioredoxin catalysis revealed by single-molecule force spectroscopy. Nat. Struct. Mol. Biol. 16, 890–896 (2009).
Carrion-Vazquez, M. et al. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA 96, 3694–3699 (1999).
Furuike, S., Ito, T. & Yamazaki, M. Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. Febs Lett. 498, 72–75 (2001).
Hutter, J.L. & Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64, 1868–1873 (1993).
Taniguchi, Y. & Kawakami, M. Application of HaloTag protein to covalent immobilization of recombinant proteins for single molecule force spectroscopy. Langmuir 26, 10433–10436 (2010).
Wang, T., Arakawa, H. & Ikai, A. Force measurement and inhibitor binding assay of monomer and engineered dimer of bovine carbonic anhydrase B. Biochem. Biophys. Res. Commun. 285, 9–14 (2001).
Kufer, S.K. et al. Covalent immobilization of recombinant fusion proteins with hAGT for single-molecule force spectroscopy. Eur. Biophys. J. Biophys. Lett. 35, 72–78 (2005).
Zakeri, B. et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 109, E690–E697 (2012).
Brujic, J., Hermans, R.I.Z., Garcia-Manyes, S., Walther, K.A. & Fernandez, J.M. Dwell-time distribution analysis of polyprotein unfolding using force-clamp spectroscopy. Biophys. J. 92, 2896–2903 (2007).
Szoszkiewicz, R. et al. Dwell time analysis of a single-molecule mechanochemical reaction. Langmuir 24, 1356–1364 (2008).
Alegre-Cebollada, J., Kosuri, P., Rivas-Pardo, J.A. & Fernandez, J.M. Direct observation of disulfide isomerization in a single protein. Nat. Chem. 3, 882–887 (2011).
Garcia-Manyes, S., Kuo, T.L. & Fernandez, J.M. Contrasting the individual reactive pathways in protein unfolding and disulfide bond reduction observed within a single protein. J. Am. Chem. Soc. 133, 3104–3113 (2011).
Bell, G.I. Models for the specific adhesion of cells to cells. Science 200, 618–627 (1978).
Evans, E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. 30, 105–128 (2001).
Dudko, O.K., Hummer, G. & Szabo, A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys. Rev. Lett. 96, 108101 (2006).
Brujic, J., Hermans, R.I., Walther, K.A. & Fernandez, J.M. Single-molecule force spectroscopy reveals signatures of glassy dynamics in the energy landscape of ubiquitin. Nat. Phys. 2, 282–286 (2006).
Berkovich, R., Garcia-Manyes, S., Urbakh, M., Klafter, J. & Fernandez, J.M. Collapse dynamics of single proteins extended by force. Biophys. J. 98, 2692–2701 (2010).
Efron, B. The Jackknife, the Bootstrap, and Other Resampling Plans (Society for Industrial and Applied Mathematics, 1982).
Kosuri, P. et al. Protein folding drives disulfide formation. Cell 151, 794–806 (2012).
Fernandez, J.M. & Li, H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science 303, 1674–1678 (2004).
Redondo-Morata, L., Giannotti, M.I. & Sanz, F. AFM-based force-clamp monitors lipid bilayer failure kinetics. Langmuir 28, 6403–6410 (2012).
Le Trong, I. et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β sheet twisting. Cell 141, 645–655 (2010).
Favre, M. et al. Force-clamp spectroscopy with a small dithering of AFM tip, and its application to explore the energy landscape of single avidin-biotin complex. Ultramicroscopy 107, 882–886 (2007).
Choy, J.L. et al. Differential force microscope for long time-scale biophysical measurements. Rev. Sci. Instrum. 78, 043711 (2007).
Zheng, P. & Li, H.B. Highly covalent ferric-thiolate bonds exhibit surprisingly low mechanical stability. J. Am. Chem. Soc. 133, 6791–6798 (2011).
Kim, J., Zhang, C.Z., Zhang, X.H. & Springer, T.A. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature 466, U992–U123 (2010).
Alegre-Cebollada, J., Badilla, C.L. & Fernandez, J.M. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J. Biol. Chem. 285, 11235–11242 (2010).
Klukovich, H.M., Kouznetsova, T.B., Kean, Z.S., Lenhardt, J.M. & Craig, S.L. A backbone lever-arm effect enhances polymer mechanochemistry. Nat. Chem. 5, 110–114 (2013).
Liu, R.C., Garcia-Manyes, S., Sarkar, A., Badilla, C.L. & Fernandez, J.M. Mechanical characterization of protein L in the low-force regime by electromagnetic tweezers/evanescent nanometry. Biophys. J. 96, 3810–3821 (2009).
Cao, Y., Kuske, R. & Li, H.B. Direct observation of Markovian behavior of the mechanical unfolding of individual proteins. Biophys. J. 95, 782–788 (2008).
Li, W.J. & Grater, F. atomistic evidence of how force dynamically regulates thiol/disulfide exchange. J. Am. Chem. Soc. 132, 16790–16795 (2010).
Zimmermann, J.L., Nicolaus, T., Neuert, G. & Blank, K. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat. Protoc. 5, 975–985 (2010).
Acknowledgements
We acknowledge all the past and present members of the Fernandez laboratory for their contribution in developing the AFM force-clamp technique. We acknowledge Luigs & Neumann for the pictures of the AFM setup and R.T. Sauer from Massachusetts Institute of Technology for the ERL-competent cells. This work was supported by grants from the US National Institutes of Health (HL066030 and HL061228 to J.M.F.). I.P. acknowledges the Swiss National Science Foundation for a postdoctoral research grant. J.A-.C. acknowledges a Fellowship from Fundación Ibercaja.
Author information
Authors and Affiliations
Contributions
All the authors performed the measurements and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Columbia University has licensed intellectual property to Luigs & Neumann GmbH.
Supplementary information
Supplementary Data
Raw data set for the mechanical unfolding of I27 in the presence of 30% glycerol, measured at four different forces. This is a single typical experiment that shows the full spectrum of challenges encountered when analyzing force-clamp measurements. (ZIP 278650 kb)
Rights and permissions
About this article
Cite this article
Popa, I., Kosuri, P., Alegre-Cebollada, J. et al. Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy. Nat Protoc 8, 1261–1276 (2013). https://doi.org/10.1038/nprot.2013.056
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.056
This article is cited by
-
Protein nanomechanics in biological context
Biophysical Reviews (2021)
-
A HaloTag-TEV genetic cassette for mechanical phenotyping of proteins from tissues
Nature Communications (2020)
-
The mechanical stability of proteins regulates their translocation rate into the cell nucleus
Nature Physics (2019)
-
Concurrent atomic force spectroscopy
Communications Physics (2019)
-
An Abl-FBP17 mechanosensing system couples local plasma membrane curvature and stress fiber remodeling during mechanoadaptation
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.