Abstract
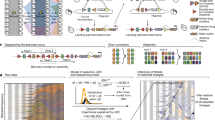
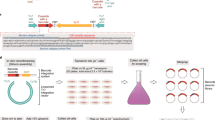
An important goal of the analysis of sequenced genomes of microbial pathogens is to improve the therapy of infectious diseases. In this context, a major challenge is to detect genomic-level evolutionary changes that increase microbial virulence. TimeZone, a genome analysis software package, is designed to detect footprints of positive selection for functionally adaptive point mutations. The uniqueness of TimeZone lies in its ability to predict recent adaptive mutations that are overlooked by conventional microevolutionary tools. This protocol describes the use of TimeZone to analyze adaptive footprints in either individual genes or in sets of genomes. Three major workflows are described: (i) extraction of orthologous gene sets from multiple genomes; (ii) alignment and phylogenetic analysis of genes; and (iii) identification of candidate genes under positive selection for point mutations, taking into account the effect of recombination events. This software package can be downloaded free from http://sourceforge.net/projects/timezone1/. In the case, for example, of the analysis of 14 Escherichia coli genomes, the protocol described here can be completed in ∼32 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Gomez-Duarte, O.G. et al. Genetic diversity of the gene cluster encoding longus, a type IV pilus of enterotoxigenic Escherichia coli. J. Bacteriol. 189, 9145–9149 (2007).
Korotkova, N. et al. Selection for functional diversity drives accumulation of point mutations in Dr adhesins of Escherichia coli. Mol. Microbiol. 64, 180–194 (2007).
Weissman, S.J. et al. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 59, 975–988 (2006).
Zhang, W. et al. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 16, 757–767 (2006).
Golding, G.B. & Dean, A.M. The structural basis of molecular adaptation. Mol. Biol. Evol. 15, 355–369 (1998).
Pallen, M.J. & Wren, B.W. Bacterial pathogenomics. Nature 449, 835–842 (2007).
Yang, Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15, 568–573 (1998).
Yang, Z. & Nielsen, R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917 (2002).
Chattopadhyay, S., Dykhuizen, D.E. & Sokurenko, E.V. ZPS: visualization of recent adaptive evolution of proteins. BMC Bioinformatics 8, 187 (2007).
Totsika, M., Beatson, S.A., Holden, N. & Gally, D.L. Regulatory interplay between pap operons in uropathogenic Escherichia coli. Mol. Microbiol. 67, 996–1011 (2008).
Weissman, S.J. et al. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl. Environ. Microbiol. 78, 1353–1360 (2012).
Dennis, G. Jr. et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3 (2003).
Sokurenko, E.V. et al. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol. Biol. Evol. 21, 1373–1383 (2004).
Chattopadhyay, S. et al. Haplotype diversity in 'source-sink' dynamics of Escherichia coli urovirulence. J. Mol. Evol. 64, 204–214 (2007).
Hughes, A.L. & Nei, M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335, 167–170 (1988).
Christin, P.-A., Weinreich, D.M. & Besnard, G. Causes and evolutionary significance of genetic convergence. Trends Genet. 26, 400–405 (2010).
Tenaillon, O. et al. The molecular diversity of adaptive convergence. Science 335, 457–461 (2012).
Chattopadhyay, S. et al. Adaptive evolution of class 5 fimbrial genes in enterotoxigenic Escherichia coli and its functional consequences. J. Biol. Chem. 287, 6150–6158 (2012).
Kisiela, D.I. et al. Convergent evolution of invasive serovars of Salmonella enterica via point mutations in the type 1 fimbrial adhesion FimH. PLoS Pathog. 8, e1002733 (2012).
Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 34, 126–129 (1992).
Weiller, G.F. Phylogenetic profiles: a graphical method for detecting genetic recombinations in homologous sequences. Mol. Biol. Evol. 15, 326–335 (1998).
Bush, R.M., Smith, C.B., Cox, N.J. & Fitch, W.M. Effects of passage history and sampling bias on phylogenetic reconstruction of human influenza A evolution. Proc. Natl Acad. Sci. USA 97, 6974–6980 (2000).
Fitch, W.M., Bush, R.M., Bender, C.A. & Cox, N.J. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl Acad. Sci. USA 94, 7712–7718 (1997).
Nei, M. & Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986).
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123, 585–595 (1989).
Fu, Y.X. & Li, W.H. Statistical tests of neutrality of mutations. Genetics 133, 693–709 (1993).
Chattopadhyay, S. et al. High frequency of hotspot mutations in core genes of Escherichia coli due to short-term positive selection. Proc. Natl Acad. Sci. USA 106, 12412–12417 (2009).
Chattopadhyay, S., Paul, S., Kisiela, D.I., Linardopoulou, E.V. & Sokurenko, E.V. Convergent molecular evolution of genomic cores in Salmonella enterica and Escherichia coli. J. Bacteriol. 194, 5002–5011 (2012).
Chattopadhyay, S., Paranjpye, R.N., Dykhuizen, D.E., Sokurenko, E.V. & Strom, M.S. Comparative evolutionary analysis of the major structural subunit of Vibrio vulnificus type IV pili. Mol. Biol. Evol. 26, 2185–2196 (2009).
Giacani, L. et al. Footprint of positive selection in Treponema pallidum subsp. pallidum genome sequences suggests adaptive microevolution of the syphilis pathogen. PLoS Negl. Trop. Dis. 6, e1698 (2012).
Paul, S. et al. Role of homologous recombination in adaptive diversification of extra-intestinal Escherichia coli. J. Bacteriol. 195, 231–242 (2013).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony and Other Methods (software) (Sinauer Associates, 2000).
Morrison, D.A. Increasing the efficiency of searches for the maximum likelihood tree in a phylogenetic analysis of up to 150 nucleotide sequences. Syst. Biol. 56, 988–1010 (2007).
Efron, B. Bootstrap methods: another look at the jackknife. Ann. Statist. 7, 1–26 (1979).
Graham, J., McNeney, B. & Seillier-Moiseiwitsch, F. Stepwise detection of recombination breakpoints in sequence alignments. Bioinformatics 21, 589–595 (2005).
Sokurenko, E.V., Gomulkiewicz, R. & Dykhuizen, D.E. Source-sink dynamics of virulence evolution. Nat. Rev. Microbiol. 4, 548–555 (2006).
Acknowledgements
We gratefully acknowledge S. Moseley (University of Washington) for critical reading of the manuscript and helpful advice. This work was supported by US National Institutes of Health (NIH) grants R01 GM084318 and RC4 AI092828.
Author information
Authors and Affiliations
Contributions
S.C. designed the protocol, wrote scripts, performed analyses and wrote the manuscript. S.P. tested and developed the protocol, and performed analyses. D.E.D. revised the manuscript. E.V.S. conceived the protocol and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Chattopadhyay, S., Paul, S., Dykhuizen, D. et al. Tracking recent adaptive evolution in microbial species using TimeZone. Nat Protoc 8, 652–665 (2013). https://doi.org/10.1038/nprot.2013.031
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.031
This article is cited by
-
Mutational convergence acts as a major player in adaptive parallel evolution of Shigella spp.
Scientific Reports (2019)
-
Recombination-independent rapid convergent evolution of the gastric pathogen Helicobacter pylori
BMC Genomics (2018)
-
Gene inversion potentiates bacterial evolvability and virulence
Nature Communications (2018)
-
A phylogeny-based sampling strategy and power calculator informs genome-wide associations study design for microbial pathogens
Genome Medicine (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.