Abstract
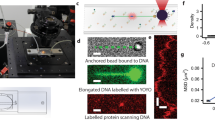
In this protocol, we describe a procedure to generate 'DNA dumbbells'—single molecules of DNA with a microscopic bead attached at each end—and techniques for manipulating individual DNA dumbbells. We also detail the design and fabrication of a microfluidic device (flow cell) used in conjunction with dual optical trapping to manipulate DNA dumbbells and to visualize individual protein-DNA complexes by single-molecule epifluorescence microscopy. Our design of the flow cell enables the rapid movement of trapped molecules between laminar flow channels and a flow-free reservoir. The reservoir provides the means to examine the formation of protein-DNA complexes in solution in the absence of external flow forces while maintaining a predetermined end-to-end extension of the DNA. These features facilitate the examination of the role of 3D DNA conformation and dynamics in protein-DNA interactions. Preparation of flow cells and reagents requires 2 days each; in situ DNA dumbbell assembly and imaging of single protein-DNA complexes require another day.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Perkins, T.T., Quake, S.R., Smith, D.E. & Chu, S. Relaxation of a single DNA molecule observed by optical microscopy. Science 264, 822–826 (1994).
Perkins, T.T., Smith, D.E. & Chu, S. Direct observation of tube-like motion of a single polymer chain. Science 264, 819–822 (1994).
Walter, N.G., Huang, C.Y., Manzo, A.J. & Sobhy, M.A. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat. Methods 5, 475–489 (2008).
Bianco, P.R. et al. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409, 374–378 (2001).
Amitani, I., Baskin, R.J. & Kowalczykowski, S.C. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell 23, 143–148 (2006).
Galletto, R., Amitani, I., Baskin, R.J. & Kowalczykowski, S.C. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443, 875–878 (2006).
Hilario, J., Amitani, I., Baskin, R.J. & Kowalczykowski, S.C. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. USA 106, 361–368 (2009).
Gross, P., Farge, G., Peterman, E.J. & Wuite, G.J. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. Methods Enzymol. 475, 427–453 (2010).
Perkins, T.T., Smith, D.E., Larson, R.G. & Chu, S. Stretching of a single tethered polymer in a uniform flow. Science 268, 83–87 (1995).
Candelli, A., Wuite, G.J. & Peterman, E.J. Combining optical trapping, fluorescence microscopy and micro-fluidics for single molecule studies of DNA-protein interactions. Phys. Chem. Chem. Phys. 13, 7263–7272 (2011).
Forget, A.L. & Kowalczykowski, S.C. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature 482, 423–427 (2012).
Brewer, L.R. & Bianco, P.R. Laminar flow cells for single-molecule studies of DNA-protein interactions. Nat. Methods 5, 517–525 (2008).
Amitani, I., Liu, B., Dombrowski, C.C., Baskin, R.J. & Kowalczykowski, S.C. Watching individual proteins acting on single molecules of DNA. Methods Enzymol. 472, 261–291 (2010).
van Mameren, J. et al. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature 457, 745–748 (2009).
van Mameren, J. et al. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl. Acad. Sci. USA 106, 18231–18236 (2009).
von Hippel, P.H. & Berg, O.G. Facilitated target location in biological systems. J. Biol. Chem. 264, 675–678 (1989).
Lomholt, M.A., van den Broek, B., Kalisch, S.M., Wuite, G.J. & Metzler, R. Facilitated diffusion with DNA coiling. Proc. Natl. Acad. Sci. USA 106, 8204–8208 (2009).
Foffano, G., Marenduzzo, D. & Orlandini, E. Facilitated diffusion on confined DNA. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 85, 021919 (2012).
van den Broek, B., Lomholt, M.A., Kalisch, S.M., Metzler, R. & Wuite, G.J. How DNA coiling enhances target localization by proteins. Proc. Natl. Acad. Sci. USA 105, 15738–15742 (2008).
Berg, O.G., Winter, R.B. & von Hippel, P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 20, 6929–6948 (1981).
Whitesides, G.M. The origins and the future of microfluidics. Nature 442, 368–373 (2006).
Lee, W.M., Reece, P.J., Marchington, R.F., Metzger, N.K. & Dholakia, K. Construction and calibration of an optical trap on a fluorescence optical microscope. Nat. Protoc. 2, 3226–3238 (2007).
Rasnik, I., Myong, S., Cheng, W., Lohman, T.M. & Ha, T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J. Mol. Biol. 336, 395–408 (2004).
Graneli, A., Yeykal, C.C., Prasad, T.K. & Greene, E.C. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir 22, 292–299 (2006).
Persson, F. et al. Lipid-based passivation in nanofluidics. Nano Lett. 12, 2260–2265 (2012).
Mirshad, J.K. & Kowalczykowski, S.C. Biochemical characterization of a mutant RecA protein altered in DNA-binding loop 1. Biochemistry 42, 5945–5954 (2003).
Harmon, F.G. & Kowalczykowski, S.C. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 12, 1134–1144 (1998).
Menetski, J.P., Bear, D.G. & Kowalczykowski, S.C. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl. Acad. Sci. USA 87, 21–25 (1990).
Acknowledgements
The authors are appreciative of members of the Kowalczykowski laboratory for their comments on this work. A.L.F. was funded by an American Cancer Society Postdoctoral Fellowship (PF-08–046–01-GMC); C.C.D. was supported by US National Institutes of Health (NIH) training grant T32 CA108459; and S.C.K. was supported by grants from the NIH (GM-62653 and GM-64745).
Author information
Authors and Affiliations
Contributions
A.L.F. and C.C.D. conceived the reservoir flow cell design and fabrication process; I.A., C.C.D. and A.L.F. built the microscope; A.L.F. and S.C.K. conceived the RecA pairing experiments; A.L.F. carried out the RecA pairing experiments and A.L.F., C.C.D., I.A. and S.C.K. wrote and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
The template used in the laser-etching step of the flow cell fabrication (PDF 2032 kb)
Rights and permissions
About this article
Cite this article
Forget, A., Dombrowski, C., Amitani, I. et al. Exploring protein-DNA interactions in 3D using in situ construction, manipulation and visualization of individual DNA dumbbells with optical traps, microfluidics and fluorescence microscopy. Nat Protoc 8, 525–538 (2013). https://doi.org/10.1038/nprot.2013.016
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.016
This article is cited by
-
A chromatinized origin reduces the mobility of ORC and MCM through interactions and spatial constraint
Nature Communications (2023)
-
Self-assembled microtubular electrodes for on-chip low-voltage electrophoretic manipulation of charged particles and macromolecules
Microsystems & Nanoengineering (2022)
-
Resonator nanophotonic standing-wave array trap for single-molecule manipulation and measurement
Nature Communications (2022)
-
Constructing artificial mimic-enzyme catalysts for carbon dioxide electroreduction
Science China Chemistry (2022)
-
DNA replication origins retain mobile licensing proteins
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.