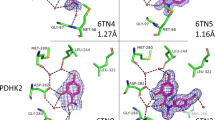
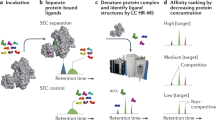
Abstract
This protocol describes the screening of a library of low-molecular-weight compounds (fragments) using a series of biophysical ligand-binding assays. Fragment-based drug discovery (FBDD) has emerged as a successful method to design high-affinity ligands for biomacromolecules of therapeutic interest. It involves detecting relatively weak interactions between the fragments and a target macromolecule using sensitive biophysical techniques. These weak binders provide a starting point for the development of inhibitors with submicromolar affinity. Here we describe an efficient fragment screening cascade that can identify binding fragments (hits) within weeks. It is divided into three stages: (i) preliminary screening using differential scanning fluorimetry (DSF), (ii) validation by NMR spectroscopy and (iii) characterization of binding fragments by isothermal titration calorimetry (ITC) and X-ray crystallography. Although this protocol is readily applicable in academic settings because of its emphasis on low cost and medium-throughput early-stage screening technologies, the core principle of orthogonal validation makes it robust enough to meet the quality standards of an industrial laboratory.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Murray, C.W. & Rees, D.C. The rise of fragment-based drug discovery. Nat. Chem. 1, 187–192 (2009).
Ciulli, A. & Abell, C. Fragment-based approaches to enzyme inhibition. Curr. Opin. Biotechnol. 18, 489–496 (2007).
Congreve, M., Carr, R., Murray, C. & Jhoti, H. A 'rule of three' for fragment-based lead discovery? Drug Discov. Today 8, 876–877 (2003).
Hung, A. et al. Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase. Angew. Chem. Int. Ed. 48, 8452–8456 (2009).
Hudson, S.A. et al. Application of fragment screening and merging to the discovery of inhibitors of the Mycobacterium tuberculosis cytochrome P450 CYP121. Angew. Chem. Int. Ed. 51, 9311–9316 (2012).
Hajduk, P.J. et al. Design of adenosine kinase inhibitors from the NMR-based screening of fragments. J. Med. Chem. 43, 4781–4786 (2000).
Gill, A., Cleasby, A. & Jhoti, H. The discovery of novel protein kinase inhibitors by using fragment-based high-throughput X-ray crystallography. Chembiochem. 6, 506–512 (2005).
Liu, G. et al. Fragment screening and assembly: a highly efficient approach to a selective and cell active protein tyrosine phosphatase 1B inhibitor. J. Med. Chem. 46, 4232–4235 (2003).
Chen, L., Cressina, E., Leeper, F.J., Smith, A.G. & Abell, C. A fragment-based approach to identifying ligands for riboswitches. ACS Chem. Biol. 5, 355–358 (2010).
Petros, A.M. et al. Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J. Med. Chem. 49, 656–663 (2006).
Scott, D.E. et al. Using a fragment-based approach to target protein-protein interactions. Chembiochem. 14, 332–342 (2013).
FDA approves vemurafenib for treatment of metastatic melanoma. Oncology 25, 906 (2011).
Baker, M. Fragment-based lead discovery grows up. Nat. Rev. Drug Discov. 12, 5–7 (2012).
Erlanson, D. (Ed.) Practical Fragments 〈http://practicalfragments.blogspot.com/〉.
Chessari, G. & Woodhead, A.J. From fragment to clinical candidate-a historical perspective. Drug Discov. Today 14, 668–675 (2009).
Scott, D.E., Coyne, A.G., Hudson, S.A. & Abell, C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry 51, 4990–5003 (2012).
Śledź, P., Abell, C. & Ciulli, A. Ligand-observed NMR in fragment-based approaches. in NMR of Biomolecules: Towards Mechanistic Systems Biology 1st edn. (eds. Bertini, I., McGreevy, K.S. & Parigi, G.), 265–281 (Wiley-VCH, 2012).
Kranz, J.K. & Schalk-Hihi, C. Protein thermal shifts to identify low molecular weight fragments. Methods Enzymol. 493, 277–298 (2011).
Hartshorn, M.J. et al. Fragment-based lead discovery using X-ray crystallography. J. Med. Chem. 48, 403–413 (2005).
Blundell, T.L., Jhoti, H. & Abell, C. High-throughput crystallography for lead discovery in drug design. Nat. Rev. Drug Discov. 1, 45–54 (2002).
Navratilova, I. & Hopkins, A.L. Fragment screening by surface plasmon resonance. ACS Med. Chem. Lett. 1, 44–48 (2010).
Hofstadler, S.A. & Sannes-Lowery, K.A. Applications of ESI-MS in drug discovery: interrogation of noncovalent complexes. Nat. Rev. Drug Discov. 5, 585–595 (2006).
Arkin, M.R. & Wells, J.A. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 3, 301–317 (2004).
Śledź, P., Lang, S., Stubbs, C.J. & Abell, C. High-throughput interrogation of ligand binding mode using a fluorescence-based assay. Angew. Chem. Int. Ed. 51, 7680–7683 (2012).
Śledź, P. et al. From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1. Angew. Chem. Int. Ed. 50, 4003–4006 (2011).
Tan, Y.S. et al. Using ligand-mapping simulations to design a ligand selectively targeting a cryptic surface pocket of polo-like kinase 1. Angew. Chem. Int. Ed. 51, 10078–10081 (2012).
Schuffenhauer, A. et al. Library design for fragment based screening. Curr. Top Med. Chem. 5, 751–762 (2005).
Siegal, G., Ab, E. & Schultz, J. Integration of fragment screening and library design. Drug Discov. Today 12, 1032–1039 (2007).
Hung, A.W. et al. Route to three-dimensional fragments using diversity-oriented synthesis. Proc. Natl. Acad. Sci. USA 108, 6799–6804 (2011).
Over, B. et al. Natural-product-derived fragments for fragment-based ligand discovery. Nat. Chem. 5, 21–28 (2013).
Tounge, B.A. & Parker, M.H. Designing a diverse high-quality library for crystallography-based FBDD screening. Methods Enzymol. 493, 3–20 (2011).
Davis, B.J. & Erlanson, D.A. Learning from our mistakes: the 'unknown knowns' in fragment screening. Bioorg. Med. Chem. Lett. 2, 2844–2852 (2013).
Alsenz, J. & Kansy, M. High throughput solubility measurement in drug discovery and development. Adv. Drug Delivery Rev. 59, 546–567 (2007).
Kozikowski, B.A. et al. The effect of freeze/thaw cycles on the stability of compounds in DMSO. J. Biomol. Screen 8, 210–215 (2003).
Poklar, N., Lah, J., Salobir, M., Macek, P. & Vesnaver, G. pH and temperature-induced molten globule-like denatured states of equinatoxin II: a study by UV-melting, DSC, far- and near-UV CD spectroscopy, and ANS fluorescence. Biochemistry 36, 14345–14352 (1997).
Pantoliano, M.W. et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen 6, 429–440 (2001).
Lo, M.-C. et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal. Biochem. 332, 153–159 (2004).
Niesen, F.H., Berglund, H. & Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 (2007).
Zhang, R. & Monsma, F. Fluorescence-based thermal shift assays. Curr. Opin. Drug Discov. Devel. 13, 389–402 (2010).
Ericsson, U.B., Hallberg, B.M., Detitta, G.T., Dekker, N. & Nordlund, P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357, 289–298 (2006).
Tjernberg, A., Markova, N., Griffiths, W.J. & Hallen, D. DMSO-related effects in protein characterization. J. Biomol. Screen 11, 131–137 (2006).
Shuker, S.B., Hajduk, P.J., Meadows, R.P. & Fesik, S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274, 1531–1534 (1996).
Lepre, C.A., Moore, J.M. & Peng, J.W. Theory and applications of NMR-based screening in pharmaceutical research. Chem. Rev. 104, 3641–3676 (2004).
Jhoti, H., Cleasby, A., Verdonk, M. & Williams, G. Fragment-based screening using X-ray crystallography and NMR spectroscopy. Curr. Opin. Chem. Biol. 11, 485–493 (2007).
Mayer, M. & Meyer, B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. 38, 1784–1788 (1999).
Dalvit, C., Fogliatto, G., Stewart, A., Veronesi, M. & Stockman, B. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J. Biomol. NMR 21, 349–359 (2001).
Carr, Y.H. & Purcell, M.E. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 94, 630–638 (1954).
Meiboom, S. & Gill, D. Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 29, 688–691 (1958).
Pellecchia, M. et al. Perspectives on NMR in drug discovery: a technique comes of age. Nat. Rev. Drug Discov. 7, 738–745 (2008).
Dalvit, C. NMR methods in fragment screening: theory and a comparison with other biophysical techniques. Drug Discov. Today 14, 1051–1057 (2009).
Angulo, J. & Nieto, P.M. STD-NMR: application to transient interactions between biomolecules-a quantitative approach. Eur. Biophys. J. 40, 1357–1369 (2011).
Dalvit, C. et al. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR 18, 65–68 (2000).
Hajduk, P.J., Olejniczak, E.T. & Fesik, S.W. One-dimensional relaxation- and diffusion-edited NMR methods for screening compounds that bind to macromolecules. J. Am. Chem. Soc. 119, 12257–12261 (1997).
Leavitt, S. & Freire, E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr. Opin. Struct. Biol. 11, 560–566 (2001).
Turnbull, W.B. & Daranas, A.H. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 125, 14859–14866 (2003).
Lewis, E.A. & Murphy, K.P. Isothermal titration calorimetry. Methods Mol. Biol. 305, 1–16 (2005).
Ladbury, J.E. Calorimetry as a tool for understanding biomolecular interactions and an aid to drug design. Biochem. Soc. Trans. 38, 888–893 (2010).
Ladbury, J.E., Klebe, G. & Freire, E. Adding calorimetric data to decision making in lead discovery: a hot tip. Nat. Rev. Drug Discov. 9, 23–27 (2010).
Śledź, P. et al. Optimization of the interligand Overhauser effect for fragment linking: application to inhibitor discovery against Mycobacterium tuberculosis pantothenate synthetase. J. Am. Chem. Soc. 132, 4544–4545 (2012).
Good, N.E. et al. Hydrogen ion buffers for biological research. Biochemistry 5, 467–477 (1966).
Hwang, T.L. & Shaka, A.J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A 112, 275–279 (1995).
McNicholas, S., Potterton, E., Wilson, K.S. & Noble, M.E.M. Presenting your structures: the CCP4mg molecular-graphics software. Acta. Crystallogr. D Biol. Crystallogr. 67, 386–394 (2011).
Acknowledgements
E.H.M. acknowledges the NIH-Cambridge Graduate Partnership Program and C. Barry (Tuberculosis Research Section, NIAID, NIH). P.Ś. thanks the Gates Cambridge Trust and St. Edmund's College for funding. S.L. thanks the German Academic Exchange Service (DAAD) and the Structural Genomics Consortium, Oxford. We thank G. Williams (Astex Pharmaceuticals) and all members of the Abell, Blundell, Hyvönen and Ciulli groups for useful discussions. This research was supported in part by the Intramural Research Program of the NIH, NIAID.
Author information
Authors and Affiliations
Contributions
C.A. initiated the project and supervised the work. E.H.M., P.Ś. and S.L. developed and formulated the protocol. E.H.M., P.Ś. and C.A. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Mashalidis, E., Śledź, P., Lang, S. et al. A three-stage biophysical screening cascade for fragment-based drug discovery. Nat Protoc 8, 2309–2324 (2013). https://doi.org/10.1038/nprot.2013.130
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.130
This article is cited by
-
MolData, a molecular benchmark for disease and target based machine learning
Journal of Cheminformatics (2022)
-
A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity
Communications Biology (2021)
-
Theory and applications of differential scanning fluorimetry in early-stage drug discovery
Biophysical Reviews (2020)
-
Fragment-based screening identifies inhibitors of ATPase activity and of hexamer formation of Cagα from the Helicobacter pylori type IV secretion system
Scientific Reports (2019)
-
Enabling STD-NMR fragment screening using stabilized native GPCR: A case study of adenosine receptor
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.