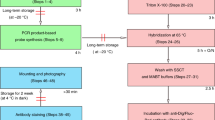
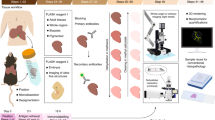
Abstract
In situ hybridization (ISH) is a powerful technique for detecting nucleic acids in cells and tissues. Here we describe three ISH procedures that are optimized for Drosophila ovaries: whole-mount, digoxigenin-labeled RNA ISH; RNA fluorescent ISH (FISH); and protein immunofluorescence (IF)–RNA FISH double labeling (IF/FISH). Each procedure balances conflicting requirements for permeabilization, fixation and preservation of antigenicity to detect RNA and protein expression with high resolution and sensitivity. The ISH protocol uses alkaline phosphatase–conjugated digoxigenin antibodies followed by a color reaction, whereas FISH detection involves tyramide signal amplification (TSA). To simultaneously preserve antigens for protein detection and enable RNA probe penetration for IF/FISH, we perform IF before FISH and use xylenes and detergents to permeabilize the tissue rather than proteinase K, which can damage the antigens. ISH and FISH take 3 d to perform, whereas IF/FISH takes 5 d. Probe generation takes 1 or 2 d to perform.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Gall, J.G. & Pardue, M.L. Formation and detection of RNA–DNA hybrid molecules in cytological preparations. Proc. Natl. Acad. Sci. USA 63, 378–383 (1969).
Pardue, M.L. & Gall, J.G. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc. Natl. Acad. Sci. USA 64, 600–604 (1969).
Tautz, D. & Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81–85 (1989).
Kallioniemi, A. et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258, 818–821 (1992).
Melton, D.A. et al. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12, 7035–7056 (1984).
Kallioniemi, A., Visakorpi, T., Karhu, R., Pinkel, D. & Kallioniemi, O.P. Gene copy number analysis by fluorescence in situ hybridization and comparative genomic hybridization. Methods 9, 113–121 (1996).
McNeil, J.A., Johnson, C.V., Carter, K.C., Singer, R.H. & Lawrence, J.B. Localizing DNA and RNA within nuclei and chromosomes by fluorescence in situ hybridization. Genet. Anal. Tech. Appl. 8, 4158 (1991).
Wiedorn, K.H., Kuhl, H., Galle, J., Caselitz, J. & Vollmer, E. Comparison of in-situ hybridization, direct and indirect in-situ PCR as well as tyramide signal amplification for the detection of HPV. Histochem. Cell Biol. 111, 89–95 (1999).
De Block, M. & Debrouwer, D. RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal. Biochem. 215, 86–89 (1993).
Harlow, E. & Lane, D. in Antibodies: A Laboratory Manual Ch. 10, 396–399 (Cold Spring Harbor Laboratory Press, 1988).
O'Neill, J.W. & Bier, E. Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. Biotechniques 17, 870, 874–875 (1994).
Yakoby, N. et al. A combinatorial code for pattern formation in Drosophila oogenesis. Dev. Cell 15, 725–737 (2008).
Lécuyer, E., Parthasarathy, N. & Krause,, H.M. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. in Drosophila: Methods and Protocols, Methods in Molecular Biology (ed. Dahmann, C.) Ch. 18, 289–302 (Humana Press, 2008).
Wilkie, G.S., Shermoen, A.W., O'Farrell, P.H. & Davis, I. Transcribed genes are localized according to chromosomal position within polarized Drosophila embryonic nuclei. Curr. Biol. 9, 1263–1266 (1999).
Fuchs, A., Cheung, L.S., Charbonnier, E., Shvartsman, S.Y. & Pyrowolakis, G. Transcriptional interpretation of the EGF receptor signaling gradient. Proc. Natl. Acad. Sci. USA 109, 1572–1577 (2012).
Kosman, D. et al. Multiplex detection of RNA expression in Drosophila embryos. Science 305, 846 (2004).
van Gijlswijk, R.P. et al. Fluorochrome-labeled tyramides: use in immunocytochemistry and fluorescence in situ hybridization. J. Histochem. Cytochem. 45, 375–382 (1997).
Spradling, A.C. Developmental genetics of oogenesis. in The Development of Drosophila melanogaster, Vol. I (eds. Bate, M. & Martinez Arias, A.) 1–70 (Cold Spring Harbor Laboratory Press, 1993).
Weiszmann, R., Hammonds, A.S. & Celniker, S.E. Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos. Nat. Protoc. 4, 605–618 (2009).
Nagaso, H., Murata, T., Day, N. & Yokoyama, K.K. Simultaneous detection of RNA and protein by in situ hybridization and immunological staining. J. Histochem. Cytochem. 49, 1177–1182 (2001).
Morris, C.A., Benson, E. & White-Cooper, H. Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nat. Protoc. 4, 1807–1819 (2009).
Toledano, H., D'Alterio, C., Loza-Coll, M. & Jones, D.L. Dual fluorescence detection of protein and RNA in Drosophila tissues. Nat. Protoc. 7, 1808–1817 (2012).
Brivanlou, A.H. & Harland, R.M. Expression of an engrailed-related protein is induced in the anterior neural ectoderm of early Xenopus embryos. Development 106, 611–617 (1989).
Harland, R.M. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36, 685–695 (1991).
Thisse, C. & Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 (2008).
Piette, D., Hendrickx, M., Willems, E., Kemp, C.R. & Leyns, L. An optimized procedure for whole-mount in situ hybridization on mouse embryos and embryoid bodies. Nat. Protoc. 3, 1194–1201 (2008).
Acloque, H., Wilkinson, D.G. & Nieto, M.A. In situ hybridization analysis of chick embryos in whole-mount and tissue sections. Methods Cell Biol. 87, 169–185 (2008).
Gillespie, D.E. & Berg, C.A. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 9, 2495–2508 (1995).
Neuman-Silberberg, F.S. & Schupbach, T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF-α–like protein. Cell 75, 165–174 (1993).
Ruohola, H. et al. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell 66, 433–449 (1991).
Ephrussi, A., Dickinson, L.K. & Lehmann, R. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66, 37–50 (1991).
Lehmann, R. & Tautz, D. In situ hybridization to RNA. in Methods in Cell Biology, Vol. 44, 575–598 (Elsevier, 1994).
Sambrook, J. & Russell, D. Molecular Cloning: A Laboratory Manual 3rd edn., Vol. 1. (Cold Spring Harbor Laboratory Press, 2001).
Schulz, C. In situ hybridization to Drosophila testes. Cold Spring Harb. Protoc. 10.1101/pdb.prot4764 (2007).
Vize, P.D., McCoy, K.E. & Zhou, X. Multichannel wholemount fluorescent and fluorescent/chromogenic in situ hybridization in Xenopus embryos. Nat. Protoc. 4, 975–983 (2009).
Wang, X. et al. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell 10, 483–495 (2006).
Pizette, S., Rabouille, C., Cohen, S.M. & Therond, P. Glycosphingolipids control the extracellular gradient of the Drosophila EGFR ligand Gurken. Development 136, 551–561 (2009).
Kitadate, Y., Shigenobu, S., Arita, K. & Kobayashi, S. Boss/Sev signaling from germline to soma restricts germline-stem-cell-niche formation in the anterior region of Drosophila male gonads. Dev. Cell 13, 151–159 (2007).
Lécuyer, E., Necakov, A.S., Caceres, L. & Krause, H.M. High-resolution fluorescent in situ hybridization of Drosophila embryos and tissues. Cold Spring Harb. Protoc. 10.1101/pdb.prot5019 (2008).
Lerner, D.W. et al. A Rab10-dependent mechanism for polarized basement membrane secretion during organ morphogenesis. Dev. Cell 24, 159–168 (2013).
Schotman, H., Karhinen, L. & Rabouille, C. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev. Cell 14, 171–182 (2008).
Vanzo, N.F. & Ephrussi, A. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development 129, 3705–3714 (2002).
Hughes, S.C. & Krause, H.M. Single and double FISH protocols for Drosophila. in Confocal Microscopy: Methods and Protocols (ed. Paddock, S.W.) Ch. 5, 93–101 (Humana Press, 1999).
Braissant, O. & Wahli, W. A simplified in situ hybridization protocol using non-radioactively labeled probes to detect abundant and rare mRNAs on tissue sections. Biochemica 1, 10–16 (1998).
Langer, P.R., Waldrop, A.A. & Ward, D.C. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc. Natl. Acad. Sci. USA 78, 6633–6637 (1981).
Peters, N.C., Thayer, N.H., Kerr, S.A., Tompa, M. & Berg, C.A. Following the 'tracks': Tramtrack69 regulates epithelial tube expansion in the Drosophila ovary through Paxillin, Dynamin, and the homeobox protein Mirror. Dev. Biol. 378, 154–169 (2013).
Geisbrecht, E.R. et al. Genetic interaction screens identify a role for Hedgehog signaling in Drosophila border cell migration. Dev. Dyn. 242, 414–431 (2013).
Kucherenko, M.M., Barth, J., Fiala, A. & Shcherbata, H.R. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 31, 4511–4523 (2012).
McLean, P.F. & Cooley, L. Protein equilibration through somatic ring canals in Drosophila. Science 340, 1445–1447 (2013).
Dernburg, A.F. Hybridization to tissues in suspension for whole-mount FISH in Drosophila. Cold Spring Harb. Protoc. 2011, 1534–1537 (2011).
Dernburg, A.F. Manual dissection and fixation of Drosophila egg chambers for whole-mount FISH. Cold Spring Harb. Protoc. 2011, 1531–1533 (2011).
Dernburg, A.F. Fragmentation and labeling of probe DNA for whole-mount FISH in Drosophila. Cold Spring Harb. Protoc. 2011, 1527–1530 (2011).
Becalska, A.N. & Gavis, E.R. Lighting up mRNA localization in Drosophila oogenesis. Development 136, 2493–2503 (2009).
Forrest, K.M. & Gavis, E.R. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 13, 1159–1168 (2003).
Randolph, J.B. & Waggoner, A.S. Stability, specificity and fluorescence brightness of multiply-labeled fluorescent DNA probes. Nucleic Acids Res. 25, 2923–2929 (1997).
Zhang, D.Y., Chen, S.X. & Yin, P. Optimizing the specificity of nucleic acid hybridization. Nat. Chem. 4, 208–214 (2012).
Femino, A.M., Fay, F.S., Fogarty, K. & Singer, R.H. Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998).
Kislauskis, E.H., Li, Z., Singer, R.H. & Taneja, K.L. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 123, 165–172 (1993).
Blanco, A.M., Rausell, L., Aguado, B., Perez-Alonso, M. & Artero, R. A FRET-based assay for characterization of alternative splicing events using peptide nucleic acid fluorescence in situ hybridization. Nucleic Acids Res. 37, e116 (2009).
Nielsen, P.E., Egholm, M., Berg, R.H. & Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254, 1497–1500 (1991).
Larsson, C., Grundberg, I., Soderberg, O. & Nilsson, M. In situ detection and genotyping of individual mRNA molecules. Nat. Methods 7, 395–397 (2010).
Nilsson, M. et al. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265, 2085–2088 (1994).
Lecuyer, E. et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 (2007).
Simin, K. et al. Profiling patterned transcripts in Drosophila embryos. Genome Res. 12, 1040–1047 (2002).
Tomancak, P. et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3, research0088.1 (2002).
Tomancak, P. et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8, R145 (2007).
Blackshaw, S. High-throughput RNA in situ hybridization in mouse retina. Methods Mol. Biol. 935, 215–226 (2013).
Chiao, E., Leonard, J., Dickinson, K. & Baker, J.C. High-throughput functional screen of mouse gastrula cDNA libraries reveals new components of endoderm and mesoderm specification. Genome Res. 15, 44–53 (2005).
Quiring, R. et al. Large-scale expression screening by automated whole-mount in situ hybridization. Mech. Dev. 121, 971–976 (2004).
King, R.C. Ovarian Development in Drosophila melanogaster (Academic Press, 1970).
Sive, H.L., Grainger, R.M. & Harland, R.M. Synthesis and purification of digoxigenin-labeled RNA probes for in situ hybridization. Cold Spring Harb. Protoc. 10.1101/pdb.prot4778 (2007).
Parrish, J.Z., Kim, M.D., Jan, L.Y. & Jan, Y.N. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 20, 820–835 (2006).
Kessler, C. Factors influencing nucleic acid hybridization. in Nonradioactive Labeling and Detection of Biomolecules (ed. Kessler, C.) Ch. 19, 253–264 (Springer, 1992).
Bodkin, D.K. & Knudson, D.L. Genetic relatedness of Palyam serogroup viruses by RNA-RNA blot hybridization. J. Gen. Virol. 67 (Part 4): 683–691 (1986).
Dubreuil, R., Byers, T.J., Branton, D., Goldstein, L.S. & Kiehart, D.P. Drosophilia spectrin. I. Characterization of the purified protein. J. Cell Biol. 105, 2095–2102 (1987).
Kim, Y. et al. Gene regulation by MAPK substrate competition. Dev. Cell 20, 880–887 (2011).
Oda, H., Uemura, T., Harada, Y., Iwai, Y. & Takeichi, M. A Drosophila homolog of Cadherin associated with Armadillo and essential for embryonic cell-cell adhesion. Dev. Biol. 165, 716–726 (1994).
Hauptmann, G. & Gerster, T. Multicolor whole-mount in situ hybridization. Methods Mol. Biol. 137, 139–148 (2000).
Acknowledgements
We thank G. Martin at the University of Washington Keck Center for imaging and advice on methods used in this protocol, P. Louie and N. Thayer for contributing to in situ hybridization optimization, T. Dodgen for a troubleshooting gel, J. Parrish and J. Lee for the use of video equipment to film ovary dissections, the Bloomington Stock Center for fly stocks and the Drosophila Genome Resources Center for cDNA clones. The α-Spectrin and E-cadherin monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, which was developed under the auspices of the National Institute for Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences. This work was supported by the University of Washington Provost Bridge Funds and NIH grant no. R01-GM079433 (C.A.B.), NIH/National Human Genome Research Institute grant no. T32 H600035 'Interdisciplinary Training in Genomic Sciences' (S.G.Z.) and National Science Foundation Graduate Research Fellowship no. DGE-0718124 (N.C.P.).
Author information
Authors and Affiliations
Contributions
C.A.B. supervised the project. C.A.B., N.C.P., A.E.A. and S.G.Z. designed the experiments. N.C.P., A.E.A. and S.G.Z. performed the experiments. C.A.B. and A.E.A. optimized the ISH methods; N.C.P., A.E.A. and S.G.Z. optimized the FISH methods; and S.G.Z. optimized the dual IF/FISH methods. N.C.P. performed dissections for Supplementary Video 1. S.G.Z. filmed and edited Supplementary Video 1. C.A.B. and S.G.Z. wrote the paper and N.C.P. and A.E.A. commented on drafts of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Video 1
Drosophila Ovary Dissection. Supplementary video showing an example of the ovary dissection procedure, including preparation, a typical dissection, dissection variations and tool maintenance. (MOV 53260 kb)
Rights and permissions
About this article
Cite this article
Zimmerman, S., Peters, N., Altaras, A. et al. Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries. Nat Protoc 8, 2158–2179 (2013). https://doi.org/10.1038/nprot.2013.136
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.136
This article is cited by
-
Loss of Paip1 causes translation reduction and induces apoptotic cell death through ISR activation and Xrp1
Cell Death Discovery (2023)
-
Molecular characterization, expression, and function of Vitellogenin genes in Phytoseiulus persimilis
Experimental and Applied Acarology (2022)
-
Improvement and use of CRISPR/Cas9 to engineer a sperm-marking strain for the invasive fruit pest Drosophila suzukii
BMC Biotechnology (2019)
-
Two Drosophilids exhibit distinct EGF pathway patterns in oogenesis
Development Genes and Evolution (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.