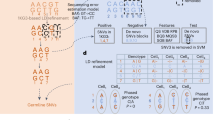
Abstract
Copy number variation (CNV) is increasingly recognized as an important contributor to phenotypic variation in health and disease. Most methods for determining CNV rely on admixtures of cells in which information regarding genetic heterogeneity is lost. Here we present a protocol that allows for the genome-wide copy number analysis of single nuclei isolated from mixed populations of cells. Single-nucleus sequencing (SNS), combines flow sorting of single nuclei on the basis of DNA content and whole-genome amplification (WGA); this is followed by next-generation sequencing to quantize genomic intervals in a genome-wide manner. Multiplexing of single cells is discussed. In addition, we outline informatic approaches that correct for biases inherent in the WGA procedure and allow for accurate determination of copy number profiles. All together, the protocol takes ∼3 d from flow cytometry to sequence-ready DNA libraries.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Accession codes
Change history
24 February 2016
In the version of this article initially published, the units for the concentration of NaCl in the NST buffer described in the Reagent Setup section were incorrect. The correct unit should be mM. The error has been corrected in the HTML and PDF versions of the article.
References
Beckmann, J.S., Estivill, X. & Antonarakis, S.E. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat. Rev. Genet. 8, 639–646 (2007).
Hasin, Y. et al. High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution. PLoS Genet. 4, e1000249 (2008).
Perry, G.H. et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39, 1256–1260 (2007).
Speliotes, E.K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007).
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010).
Bignell, G.R. et al. Signatures of mutation and selection in the cancer genome. Nature 463, 893–898 (2010).
Russnes, H.G. et al. Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Sci. Transl. Med. 2, 38ra47 (2010).
Shiu, K.K., Natrajan, R., Geyer, F.C., Ashworth, A. & Reis-Filho, J.S. DNA amplifications in breast cancer: genotypic-phenotypic correlations. Future Oncol. 6, 967–984 (2010).
Shinawi, M. & Cheung, S.W. The array CGH and its clinical applications. Drug Discov. Today 13, 760–770 (2008).
Santarius, T., Shipley, J., Brewer, D., Stratton, M.R. & Cooper, C.S. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 10, 59–64 (2010).
Pinkel, D. & Albertson, D.G. Array comparative genomic hybridization and its applications in cancer. Nat. Genet. 37 (suppl.): S11–17 (2005).
Praulich, I. et al. Clonal heterogeneity in childhood myelodysplastic syndromes—challenge for the detection of chromosomal imbalances by array-CGH. Genes Chromosomes Cancer 49, 885–900 (2010).
Metzker, M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 11, 31–46 (2010).
Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133–141 (2008).
Meyerson, M., Gabriel, S. & Getz, G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696 (2010).
Chiang, D.Y. et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods 6, 99–103 (2009).
Alkan, C. et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat. Genet. 41, 1061–1067 (2009).
Shah, S.P. et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 (2009).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Campbell, P.J. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 15, 2078–2079 (2009).
Venkatraman, E.S. & Olshen, A.B. A faster cicular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23, 657–63 (2007).
Wersto, R.P. et al. Doublet discrimination in DNA cell-cycle analysis. Cytometry 46, 296–306 (2001).
Nagata, S., Nagase, H., Kawane, K., Mukae, N. & Fukuyama, H. Degradation of chromosomal DNA during apoptosis. Cell Death Differ 10, 108–116 (2003).
Stephans, P.J. et al. Massive genomic rearragement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011).
Acknowledgements
We acknowledge all the members of the Hicks and Wigler labs for discussions during the technology development. We thank W. McCombie, E. Ghiban and L. Gelley for their advice and technical assistance, and A. Gordon for informatics support and assistance with figures. N.N. is supported by grants from Texas STARS and the Alice Kleberg Reynolds Foundation. This work was supported by grants to M.W. and J.H. from the Department of the Army (W81XWH04-1-0477), the Breast Cancer Research Foundation, and the Simons Foundation. M.W. is an American Cancer Society Research Professor.
Author information
Authors and Affiliations
Contributions
T.B., J.K., N.N., J.H. and M.W. designed methods and analyzed data. T.B., L.R., H.C., M.R., A.S. and J.T. performed experiments. K.R., D.E. and B.L. contributed to the technology development. J.K., N.N., T.B., J.H. and M.W. developed informatic analysis. T.B., J.K. and J.H. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Outline of the informatics section. Provide a concise outline of all the steps with input-program-output labels. (EPS 1481 kb)
Supplementary Data
Input/output data for the informatics procedure of the protocol required to reproduce copy number profiles from single cell sequencing data. Supplementary Data Legend outlines all the data with descriptions. (ZIP 78479 kb)
Supplementary Methods
Contains all programs required to process single cell sequencing data for copy number determination, from genome preparation to sequence mapping and copy number estimation. Supplementary Methods Legend outlines all the programs and their utilities. (ZIP 107 kb)
Supplementary Note
Descriptions of data and program files. Text providing a brief overview of the data and program files included in the Supplementary Methods and Supplementary Data. (TXT 2 kb)
Rights and permissions
About this article
Cite this article
Baslan, T., Kendall, J., Rodgers, L. et al. Genome-wide copy number analysis of single cells. Nat Protoc 7, 1024–1041 (2012). https://doi.org/10.1038/nprot.2012.039
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.039
This article is cited by
-
The reckoning of chromosomal instability: past, present, future
Chromosome Research (2024)
-
COMPASS: joint copy number and mutation phylogeny reconstruction from amplicon single-cell sequencing data
Nature Communications (2023)
-
Chromosome instability and aneuploidy in the mammalian brain
Chromosome Research (2023)
-
HBV genome-enriched single cell sequencing revealed heterogeneity in HBV-driven hepatocellular carcinoma (HCC)
BMC Medical Genomics (2022)
-
Somatic copy number variant load in neurons of healthy controls and Alzheimer’s disease patients
Acta Neuropathologica Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.