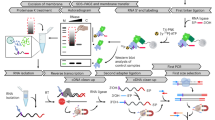
Abstract
Owing to the low abundance of signaling proteins and transcription factors, their protein complexes are not easily identified by classical proteomics. The isolation of these protein complexes from endogenous plant tissues (rather than plant cell cultures) is therefore an important technical challenge. Here, we describe a sensitive, quantitative proteomics-based procedure to determine the composition of plant protein complexes. The method makes use of fluorophore-tagged protein immunoprecipitation (IP) and label-free mass spectrometry (MS)-based quantification to correct for nonspecifically precipitated proteins. We provide procedures for the isolation of membrane-bound receptor complexes and transcriptional regulators from nuclei. The protocol consists of an IP step (∼6 h) and sample preparation for liquid chromatography-tandem MS (LC-MS/MS; 2 d). We also provide a guide for data analysis. Our single-step affinity purification protocol is a good alternative to two-step tandem affinity purification (TAP), as it is shorter and relatively easy to perform. The data analysis by label-free quantification (LFQ) requires a cheaper and less challenging experimental setup compared with known labeling techniques in plants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rohila, J.S., Chen, M., Cerny, R. & Fromm, M.E. Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38, 172–181 (2004).
Rohila, J.S. et al. Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 46, 1–13 (2006).
Rubio, V. et al. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778 (2005).
Van Leene, J. et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol. Syst. Biol. 6, 397 (2010).
Blagoev, B. et al. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 (2003).
Rinner, O. et al. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat. Biotechnol. 25, 345–352 (2007).
Hubner, N.C. et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–754 (2010).
America, A.H. & Cordewener, J.H. Comparative LC-MS: a landscape of peaks and valleys. Proteomics 8, 731–749 (2008).
Clough, S.J. & Bent, A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Gruhler, A., Schulze, W.X., Matthiesen, R., Mann, M. & Jensen, O.N. Stable isotope labeling of Arabidopsis thaliana cells and quantitative proteomics by mass spectrometry. Mol. Cell. Proteomics 4, 1697–1709 (2005).
Poser, I. et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods 5, 409–415 (2008).
Smaczniak, C. et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109, 1560–1565 (2012).
Theissen, G. & Saedler, H. Plant biology. Floral quartets. Nature 409, 469–471 (2001).
Mravec, J. et al. Cell plate restricted association of DRP1A and PIN proteins is required for cell polarity establishment in Arabidopsis. Curr. Biol. 21, 1055–1060 (2011).
Gudesblat, G.E. et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548–554 (2012).
Karlova, R. et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18, 626–638 (2006).
Karlova, R. The SERK1 Protein Complexes Ph.D thesis, Wageningen University (2008).
Immink, R.G.H., Kaufmann, K. & Angenent, G.C. The 'ABC' of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol. 21, 87–93 (2010).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Zhu, W., Smith, J.W. & Huang, C.M. Mass spectrometry-based label-free quantitative proteomics. J. Biomed. Biotechnol. 2010, 840518 (2010).
Ning, K., Fermin, D. & Nesvizhskii, A.I. Comparative analysis of different label-free mass spectrometry based protein abundance estimates and their correlation with RNA-Seq gene expression data. J. Proteome Res. 11, 2261–2271 (2012).
Old, W.M. et al. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell Proteomics 4, 1487–1502 (2005).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Deal, R.B. & Henikoff, S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 6, 56–68 (2011).
Melzer, R., Verelst, W. & Theissen, G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in 'floral quartet'-like complexes in vitro. Nucleic Acids Res. 37, 144–157 (2009).
Schwanhausser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Curtis, M.D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Hecht, V. et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816 (2001).
Kwaaitaal, M.A., de Vries, S.C. & Russinova, E. Arabidopsis thaliana Somatic Embryogenesis Receptor Kinase 1 protein is present in sporophytic and gametophytic cells and undergoes endocytosis. Protoplasma 226, 55–65 (2005).
Savitski, M.M., Nielsen, M.L. & Zubarev, R.A. ModifiComb: New proteomics tool for mapping substochiometric post-translational modifications, finding novel types of modifications and fingerprinting complex protein mixtures. Mol. Cell Proteomics 5, 935–948 (2006).
Palmisano, G., Melo-Braga, M.N., Engholm-Keller, K., Parker, B.L. & Larsen, M.R. Chemical Deamidation: a common pitfall in large-scale N-linked glycoproteomic mass spectrometry-based analyses. J. Proteome Res. 11, 1949–1957 (2012).
Kersey, P.J. et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 (2004).
Cox, J. et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 (2009).
Zhou, M. & Veenstra, T.D. Proteomic analysis of protein complexes. Proteomics 7, 2688–2697 (2007).
Kaufmann, K., Smaczniak, C., de Vries, S., Angenent, G.C. & Karlova, R. Proteomics insights into plant signaling and development. Proteomics 11, 744–755 (2011).
Berggard, T., Linse, S. & James, P. Methods for the detection and analysis of protein-protein interactions. Proteomics 7, 2833–2842 (2007).
Dunham, W.H., Mullin, M. & Gingras, A.C. Affinity-purification coupled to mass spectrometry: Basic principles and strategies. Proteomics 12, 1576–1590 (2012).
Pflieger, D., Bigeard, J. & Hirt, H. Isolation and characterization of plant protein complexes by mass spectrometry. Proteomics 11, 1824–1833 (2011).
Acknowledgements
C.S. and G.C.A. were supported by a Proteomics Hotel grant from the Netherlands Proteomics Centre (NPCII-T2.3) and Centre for BioSystems Genomics (CBSG2012). All proteomic measurements were recorded at Biqualys Wageningen (http://www.biqualys.nl). N.L. was supported by the Netherlands Organization for Science, ALW grant no. 61.62.5000.86. N.L., W.v.D. and S.d.V. thank B. Möller for sharing details of IP protocols and helpful discussions. K.K. thanks M.C. O'Flaherty and R. Karlova for advice during initial stages of the project, as well as J.H. Cordewener and J.J.M. Vervoort for useful discussions. We thank J. Muiño for support in bioinformatics and statistical analysis.
Author information
Authors and Affiliations
Contributions
C.S., N.L. and K.K. designed the research; C.S., N.L. and W.v.D. performed experiments; S.B. recorded LC-MS/MS measurements; C.S. and S.B. analyzed the data; S.B., T.A., S.d.V. and G.C.A. commented on the manuscript; S.S.G. provided trypsin digestion and peptide purification protocols for soluble proteins; K.K. supervised the project; C.S., N.L. and K.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Methods
Methods for Label-free quantification with Progenesis LC-MS; Bimolecular fluorescence complementation (BiFC) and Fluorescence Resonance Energy Transfer-Fluorescence Lifetime Imaging Microscopy (FRET-FLIM). (PDF 274 kb)
Supplementary Note
A small database of sequences for background proteins that we expect to find in all samples. These are present in FASTA format to perform the first search. (TXT 32 kb)
Supplementary Data 1
MaxQuant analysis results of the PI-GFP and Col-0 immunoprecipitates: (A) MaxQuant output (proteinGroups.txt) dataset of protein quantification; (B) Statistical analysis with Perseus of the proteinGroups.txt dataset according to Steps 53-62 of the protocol; (C) Input parameters for MaxQuant; (D) Experimental design input table. The results presented here are based on the same raw data used in REF. 11. (XLSX 368 kb)
Supplementary Data 2
MaxQuant analysis results of the SERK1-CFP and Col-0 immunoprecipitates: (A) MaxQuant output (proteinGroups.txt) dataset of protein quantification; (B) Statistical analysis with Perseus of the proteinGroups.txt dataset according to Steps 53-62 of the protocol; (C) Input parameters for MaxQuant; (D) Experimental design input table. (XLSX 302 kb)
Supplementary Figure 1
Protein interaction profiling using Progenesis LC-MS on the example of PI-GFP IPs. Graphical representation of the normalized protein abundance ratios between the PI-GFP IP samples and controls plotted against total protein normalized abundance (summed peptide MS1 peak abundances – areas under peak). Imputation of the missing values with the lowest normalized protein abundance value. (PDF 938 kb)
Supplementary Figure 2
Interactions of SERK1 with At1g27190, At2g41820 and At3g28450 confirmed by FRET-FLIM and BiFC in Arabidopsis protoplasts. (A-C) Fluorescence lifetime images of A. thaliana leaf protoplasts transiently expressing SERK1-sCFP (A), SERK1-sCFP + At2g41820-sYFP (B) and SERK1-sCFP + At1g27190-sYFP (C) for 16 hrs. sCFP lifetime distributions are presented as pseudocolor images; the color bar shows the lifetime distribution, ranging from t = 2.0 ns (red) – 3.0 ns (blue). Average lifetimes, determined on at least 45 protoplasts in three independent experiments, are 2.62 ± 0.06 ns for SERK1-sCFP, 2.37 ± 0.06 ns for SERK1-sCFP/At1g27190-sYFP and 2.46 ± 0.09ns for SERK1-sCFP/At2g41820-sYFP. (D-F) Bimolecular fluorescence complementation of SERK1-YFPC with At1g27190-YFPN (D), At2g41820-YFPN (E) and At3g28450-YFPN (F) expressed in Arabidopsis leaf protoplasts for 16 hrs. (PDF 1516 kb)
Rights and permissions
About this article
Cite this article
Smaczniak, C., Li, N., Boeren, S. et al. Proteomics-based identification of low-abundance signaling and regulatory protein complexes in native plant tissues. Nat Protoc 7, 2144–2158 (2012). https://doi.org/10.1038/nprot.2012.129
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.129
This article is cited by
-
Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro
Scientific Reports (2020)
-
Dynamic and spatial restriction of Polycomb activity by plant histone demethylases
Nature Plants (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.